
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,607,058 – 15,607,320 |
| Length | 262 |
| Max. P | 0.831322 |

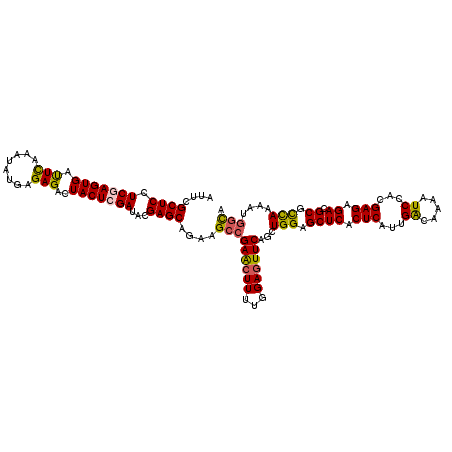
| Location | 15,607,058 – 15,607,178 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -27.10 |
| Energy contribution | -26.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
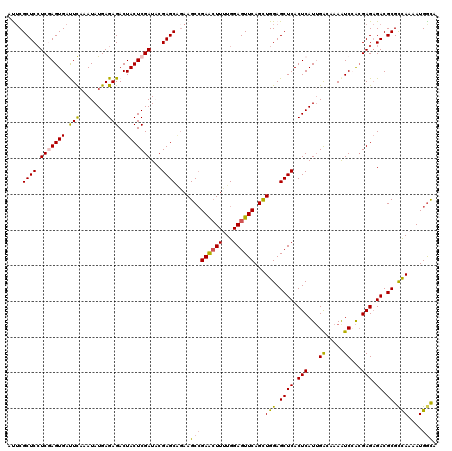
>X_DroMel_CAF1 15607058 120 + 22224390 AUUCGCUCAUCGAGUGAUUCAAAUAUGAGAGACUACUCGAUACGAGCAGAAGCCGAACUUUUGGAGUUCAGCUGGAGCUCACUCAUUGGCAAAAUCCAAGAGAGACGCGCCAAAAUGGCA .(((((((((((((((.(((........)))..))))))))..)))).)))(((((((((...))))))...(((.((((.(((.((((......))))))).)).)).)))....))). ( -40.10) >DroSec_CAF1 5137 120 + 1 AUUCGCUCCUCGAGUGACUCAAAUAUGAGAGACUACUCGAUCCGAGCUGAAGCCGAACUUCUGGAGUUCAGCUGGAGCUCACUCAUUGACAAAAUCCACGAGAGACGCGCCAAAAUGGCA ....(((((((((((..((((....))))..)))..(((((..(((((..(((.((((((...)))))).)))..)))))....))))).........)))).)).))(((.....))). ( -38.40) >DroSim_CAF1 5093 120 + 1 AUUCGCUCCUCGAGUGACUCAAAUAUGAGAGACUACUCGAUCCGAGCGGAAGCCGAACUUCUGGAGUUCAGCUGGAGCUCACUCAUUGACAAAAUCCACGAGAGACGCGCCAAAAUGACA .(((((((.(((((((.(((........)))..)))))))...)))))))(((.((((((...)))))).)))((.((((.(((..((........)).))).)).)).))......... ( -36.80) >DroEre_CAF1 5138 120 + 1 ACUCGCUCCUCAAGUGAUUUAAAUAUGAGAGACUACUCGAUACGAGCACAAGCGGAAAUUUUGGAGUUCAGUUGGAGCUCACUCAUAGACAAAAUCGGCGAGAGACGCGUUAAAAUGGCA ...(((..(((..(((((((...((((((......((((...)))).................((((((.....)))))).))))))....))))).))..)))..)))........... ( -27.60) >DroYak_CAF1 5115 120 + 1 ACACGCUCCUCAAGUGAUUUUAAUAUGAGAGACUACUCGAUACGAGCGGAAGCCGAGCUUUUGGAGUUCAGUUGGAGCUCACUCAUUGACAAAGUCAACGAGCGACGCGCUAAAAUGGUC ...((((((((...(((((((...(((((......((((...))))(((((((...)))))))((((((.....)))))).)))))....)))))))..))).)).)))........... ( -33.70) >consensus AUUCGCUCCUCGAGUGAUUCAAAUAUGAGAGACUACUCGAUACGAGCAGAAGCCGAACUUUUGGAGUUCAGCUGGAGCUCACUCAUUGACAAAAUCCACGAGAGACGCGCCAAAAUGGCA ....((((.(((((((.(((........)))..)))))))...))))....(((((((((...))))))...(((.((((.(((...((.....))...))).)).)).)))....))). (-27.10 = -26.74 + -0.36)

| Location | 15,607,058 – 15,607,178 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -28.40 |
| Energy contribution | -29.44 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15607058 120 - 22224390 UGCCAUUUUGGCGCGUCUCUCUUGGAUUUUGCCAAUGAGUGAGCUCCAGCUGAACUCCAAAAGUUCGGCUUCUGCUCGUAUCGAGUAGUCUCUCAUAUUUGAAUCACUCGAUGAGCGAAU .(((.....)))((.((.(((((((......)))).))).))))...(((((((((.....)))))))))..(((((..(((((((.(..((........))..)))))))))))))... ( -41.70) >DroSec_CAF1 5137 120 - 1 UGCCAUUUUGGCGCGUCUCUCGUGGAUUUUGUCAAUGAGUGAGCUCCAGCUGAACUCCAGAAGUUCGGCUUCAGCUCGGAUCGAGUAGUCUCUCAUAUUUGAGUCACUCGAGGAGCGAAU .(((.....))).(((..((((.((((((.....(((((.(((((..(((((((((.....)))))))))..)))))((((......)))))))))....))))).).))))..)))... ( -45.20) >DroSim_CAF1 5093 120 - 1 UGUCAUUUUGGCGCGUCUCUCGUGGAUUUUGUCAAUGAGUGAGCUCCAGCUGAACUCCAGAAGUUCGGCUUCCGCUCGGAUCGAGUAGUCUCUCAUAUUUGAGUCACUCGAGGAGCGAAU ........(((.((.((.(((((.(((...))).))))).)))).)))((((((((.....))))))))...(((((...((((((.(.(((........))).))))))).)))))... ( -45.40) >DroEre_CAF1 5138 120 - 1 UGCCAUUUUAACGCGUCUCUCGCCGAUUUUGUCUAUGAGUGAGCUCCAACUGAACUCCAAAAUUUCCGCUUGUGCUCGUAUCGAGUAGUCUCUCAUAUUUAAAUCACUUGAGGAGCGAGU ...........(((..(((..(..(((((....((((((.(((.((.....)).)))...............((((((...))))))....))))))...))))).)..)))..)))... ( -25.00) >DroYak_CAF1 5115 120 - 1 GACCAUUUUAGCGCGUCGCUCGUUGACUUUGUCAAUGAGUGAGCUCCAACUGAACUCCAAAAGCUCGGCUUCCGCUCGUAUCGAGUAGUCUCUCAUAUUAAAAUCACUUGAGGAGCGUGU ..........((((.((((((((((((...))))))))))))(((((....((.((.....)).))((((...(((((...))))))))).....................))))))))) ( -36.80) >consensus UGCCAUUUUGGCGCGUCUCUCGUGGAUUUUGUCAAUGAGUGAGCUCCAGCUGAACUCCAAAAGUUCGGCUUCCGCUCGUAUCGAGUAGUCUCUCAUAUUUGAAUCACUCGAGGAGCGAAU ...........(((.((.((((..(((((.....((((.(((((...(((((((((.....)))))))))...)))))....(((....)))))))....)))))...)))))))))... (-28.40 = -29.44 + 1.04)

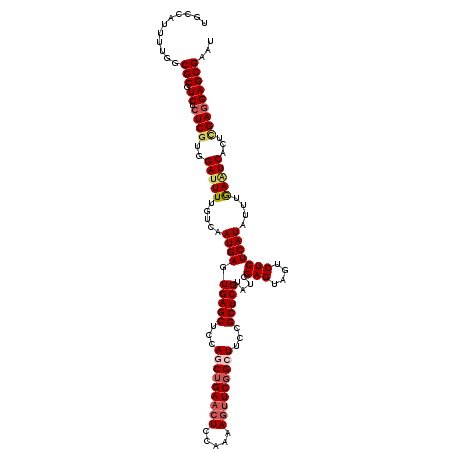
| Location | 15,607,178 – 15,607,280 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.09 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -14.86 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
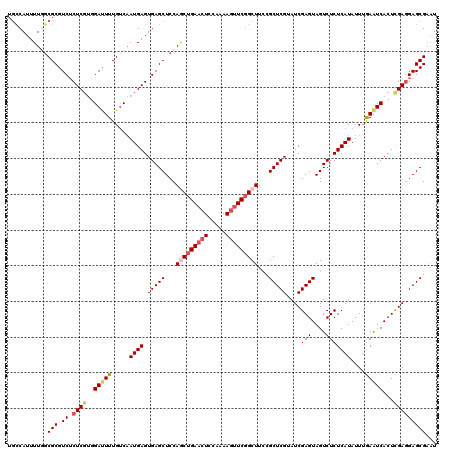
>X_DroMel_CAF1 15607178 102 - 22224390 ACGUUUGUUGUAAAACUACAAGUGGCGGAAAUGAAGUGAAUGAUAUGAGCCAUUUCAAUAUCUAAGAUAUUAG---U---------------ACGGUUCUACUUACUUUCGCUGCUUACU ..((((......))))...(((..((((((...(((((........(((((.....((((((...))))))..---.---------------..))))))))))..))))))..)))... ( -24.00) >DroSec_CAF1 5257 105 - 1 ACGUUUGUUGUAAAACUACAAGUGGCGGAAAUGGAGUGAAUGAUAUGAGCCACUUCCAUAUCUAAGAUAUCAGAUAU---------------UAGGUUCUACUUACUUUCGCUGCUUACU ..((((......))))...(((..((((((...((((((((((((((.........))))))...(((((...))))---------------)..)))).))))..))))))..)))... ( -24.30) >DroSim_CAF1 5213 105 - 1 ACGUUCGUUGUAAAACUACAAGUGGCGGAAAUGGAGUGAACGAUAUGAGCCAUUUCCAUAUCUAAGAUAUUAGAUAU---------------UAGGUUCUACUUACUUUCGCUGCUUACU ..((..(((....))).))(((..((((((...((((((((((.(((...))).)).((((((((....))))))))---------------...)))).))))..))))))..)))... ( -27.00) >DroEre_CAF1 5258 102 - 1 ACGUUUGUUGUGAAACUGAAAGUAGCGGAAAUAAAGUGAA--------AUG------AAUUGAAAGUUAUGAG---UGGUA-UUGUAAUUACAGUGUUCUACUUACUUUCGCUGCUUAUU ..((((......))))...(((((((((((...(((((((--------...------.((((..(((((..(.---.....-)..))))).)))).))).))))..)))))))))))... ( -23.30) >DroYak_CAF1 5235 114 - 1 ACGUUGGUUGUAAAACUGCAAGUAGCGUAAAUGAAGUGAACGAUAUUAAAG------AUAUGAAAGAUAUGAGAUAUGAUAGAUAUAAUUACAUUGUUCCACUCACUUUCGCUGCUUGUU .....((((....))))((((((((((.((.(((.(((((((((.......------((((.....))))...((((.....))))......)))))).)))))).)).)))))))))). ( -25.20) >consensus ACGUUUGUUGUAAAACUACAAGUGGCGGAAAUGAAGUGAACGAUAUGAGCCA_UUC_AUAUCUAAGAUAUGAGAUAU_______________AAGGUUCUACUUACUUUCGCUGCUUACU ......(((....)))...(((((((((((...((((((((......................................................)))).))))..)))))))))))... (-14.86 = -14.90 + 0.04)


| Location | 15,607,206 – 15,607,320 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.51 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15607206 114 + 22224390 ---A---CUAAUAUCUUAGAUAUUGAAAUGGCUCAUAUCAUUCACUUCAUUUCCGCCACUUGUAGUUUUACAACAAACGUAUUCAUUCCGGAUAUUCGAGUGGCAUUCGCACCCAACUCA ---.---..((((((...))))))(((((((...............))))))).((((((((..((((......))))((((((.....)))))).))))))))................ ( -21.36) >DroSec_CAF1 5285 117 + 1 ---AUAUCUGAUAUCUUAGAUAUGGAAGUGGCUCAUAUCAUUCACUCCAUUUCCGCCACUUGUAGUUUUACAACAAACGUAUUCAUUCCGGACAUUCGAGUGGCAUUCGCACCCAACUCA ---((((((((....))))))))((((((((...............))))))))((((((((..(((((((.......)))........))))...))))))))................ ( -28.06) >DroSim_CAF1 5241 117 + 1 ---AUAUCUAAUAUCUUAGAUAUGGAAAUGGCUCAUAUCGUUCACUCCAUUUCCGCCACUUGUAGUUUUACAACGAACGUAUUCAUUCCGGACAUUCGAGUGGCAUUCGCACCCAACUCA ---((((((((....))))))))((((((((...............))))))))((((((((..((((......(((....))).....))))...))))))))................ ( -28.56) >DroYak_CAF1 5275 114 + 1 AUCAUAUCUCAUAUCUUUCAUAU------CUUUAAUAUCGUUCACUUCAUUUACGCUACUUGCAGUUUUACAACCAACGUAUUCAUACCGAACAUCCAAGUGGCAUUCGCGCCCAACUCA .......................------.........................((((((((..(((((((.......)))........))))...))))))))................ ( -11.30) >consensus ___AUAUCUAAUAUCUUAGAUAUGGAAAUGGCUCAUAUCAUUCACUCCAUUUCCGCCACUUGUAGUUUUACAACAAACGUAUUCAUUCCGGACAUUCGAGUGGCAUUCGCACCCAACUCA ..................(((((((.......)))))))...............((((((((..((((......))))...(((.....)))....))))))))................ (-15.31 = -15.88 + 0.56)

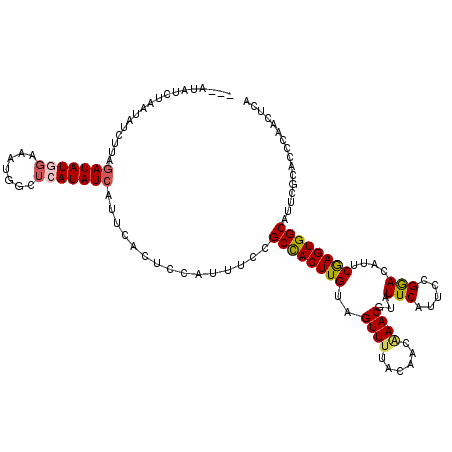
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:34 2006