
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,515,241 – 15,515,382 |
| Length | 141 |
| Max. P | 0.979891 |

| Location | 15,515,241 – 15,515,351 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15515241 110 + 22224390 AAAAAUGGCAUGGGAAGGAACAUUGAAUGGCCAAAAAUUAGCAUCUUUUACAUUCGAAAAUGUCAAAAGAAGAUGCAUCA-AAGCGGCGGCAUGAUUUCAAUAUAAUCCCC ...........((((......((((((..(((........((((((((((((((....)))))....))))))))).((.-....)).))).....))))))....)))). ( -23.60) >DroSec_CAF1 75703 97 + 1 ---------AUG----GGAACCUUAAACCGCCAGAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCA-AGGCGGCGGCAUGAUUUCAAUAUAAUCCCA ---------.((----(((........(((((........(((((((((.((((....)))).....)))))))))....-....)))))..((....))......))))) ( -26.89) >DroSim_CAF1 87687 98 + 1 ---------AUG----GGAACCUUAAACCGCCAAAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCGGCGGCGGCGGCAUGAUUUCAAUAUAAUCCCA ---------.((----(((........(((((........(((((((((.((((....)))).....)))))))))...(....))))))..((....))......))))) ( -27.00) >consensus _________AUG____GGAACCUUAAACCGCCAAAAAUUAGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCA_AGGCGGCGGCAUGAUUUCAAUAUAAUCCCA ................(((........(((((........(((((((((.((((....)))).....))))))))).........)))))..((....))......))).. (-19.14 = -19.03 + -0.11)

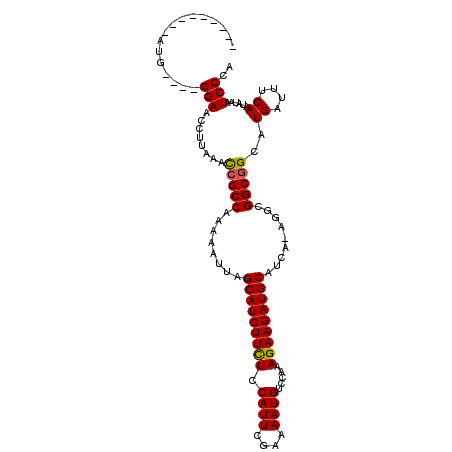
| Location | 15,515,241 – 15,515,351 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.70 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.59 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15515241 110 - 22224390 GGGGAUUAUAUUGAAAUCAUGCCGCCGCUU-UGAUGCAUCUUCUUUUGACAUUUUCGAAUGUAAAAGAUGCUAAUUUUUGGCCAUUCAAUGUUCCUUCCCAUGCCAUUUUU (((((..(((((((....(((..((((...-....(((((((......(((((....)))))..))))))).......))))))))))))))...)))))........... ( -26.64) >DroSec_CAF1 75703 97 - 1 UGGGAUUAUAUUGAAAUCAUGCCGCCGCCU-UGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUCUGGCGGUUUAAGGUUCC----CAU--------- (((((......((....)).(((((((((.-.((.(((((((((..(..(......)..)..))))))))).....)).))))))....))))))----)).--------- ( -33.40) >DroSim_CAF1 87687 98 - 1 UGGGAUUAUAUUGAAAUCAUGCCGCCGCCGCCGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUUUGGCGGUUUAAGGUUCC----CAU--------- (((((......((....)).(((...((((((((.(((((((((..(..(......)..)..)))))))))......))))))))....))))))----)).--------- ( -36.00) >consensus UGGGAUUAUAUUGAAAUCAUGCCGCCGCCU_UGAUGCAUCUUCUUUGGACAUUUUCGAAUGGAGAAGAUGCUAAUUUUUGGCGGUUUAAGGUUCC____CAU_________ .(((((....((((......(((((((........((((((((((....((((....)))))))))))))).......))))))))))).)))))................ (-22.48 = -23.59 + 1.11)

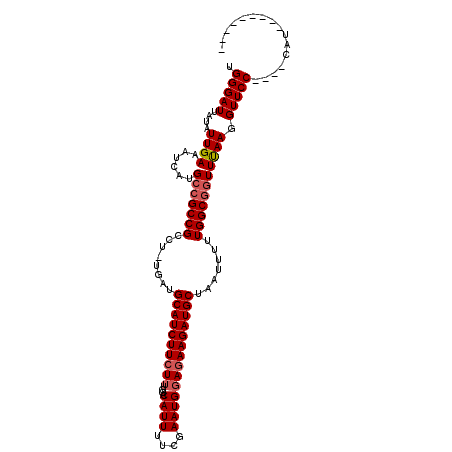

| Location | 15,515,280 – 15,515,382 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15515280 102 + 22224390 AGCAUCUUUUACAUUCGAAAAUGUCAAAAGAAGAUGCAUCA-AAGCGGCGGCAUGAUUUCAAUAUAAUCCCCGAGUGCCACGAAAAUAAUCAAUCAAUUUCUU .((((((((((((((....)))))....)))))))))....-...((..(((((((((.......)))).....))))).))..................... ( -20.20) >DroSec_CAF1 75729 102 + 1 AGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCA-AGGCGGCGGCAUGAUUUCAAUAUAAUCCCAGAAUGCGCCGAAAAUAAUCAAUCAAUUUCUU .(((((((((.((((....)))).....)))))))))....-...(((((.(((((((.......)))).....))))))))..................... ( -22.80) >DroSim_CAF1 87713 103 + 1 AGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCGGCGGCGGCGGCAUGAUUUCAAUAUAAUCCCAGAAUGCGCCGAAAAUAAUCAAUCAAUUUCUU .(((((((((.((((....)))).....))))))))).((((((((....))..((((.......))))........)))))).................... ( -25.00) >consensus AGCAUCUUCUCCAUUCGAAAAUGUCCAAAGAAGAUGCAUCA_AGGCGGCGGCAUGAUUUCAAUAUAAUCCCAGAAUGCGCCGAAAAUAAUCAAUCAAUUUCUU .(((((((((.((((....)))).....)))))))))........(((((.(((((((.......)))).....))))))))..................... (-18.77 = -19.00 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:52 2006