
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,514,063 – 15,514,254 |
| Length | 191 |
| Max. P | 0.980261 |

| Location | 15,514,063 – 15,514,162 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.83 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514063 99 + 22224390 CUGGCGAUCUGAUCGAUCGGCGGCUGAACAGUUGCAAAAACAGCCA----GCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC------- (((((((((.....)))).((((((....)))))).......))))----).........((((((((((.((((..(....)..))))..)).)))))))).------- ( -38.20) >DroSec_CAF1 74525 99 + 1 CUGGCGAUCUGAUCGAUCGGCGGCUGAACAGUUGCAAAAACAGCCA----GCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC------- (((((((((.....)))).((((((....)))))).......))))----).........((((((((((.((((..(....)..))))..)).)))))))).------- ( -38.20) >DroSim_CAF1 86531 103 + 1 CUGGCGAUCUGAUCGAUCGGCGGCUGAACAGUUGCAAAAACAGCCA----GCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC---UUUU (((((((((.....)))).((((((....)))))).......))))----).........((((((((((.((((..(....)..))))..)).)))))))).---.... ( -38.20) >DroEre_CAF1 76412 103 + 1 CUGGCGAUCUGAUCGAUCGGCGGCUGAACAGUUGCAAAAACAGCCGAGGAAC-------CGGGAAACCCCGAAGGACCCAGCGAACCUGAGGGCGGGUUGAGCGGCUGAG ....(((((.....)))))((((((....)))))).....((((((.((..(-------((((....))))..)..))).((.((((((....))))))..))))))).. ( -39.40) >consensus CUGGCGAUCUGAUCGAUCGGCGGCUGAACAGUUGCAAAAACAGCCA____GCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC_______ .((((((((.....)))).((((((....)))))).......))))..............((((((((((.((((..........))))..)).))))))))........ (-29.76 = -30.83 + 1.06)

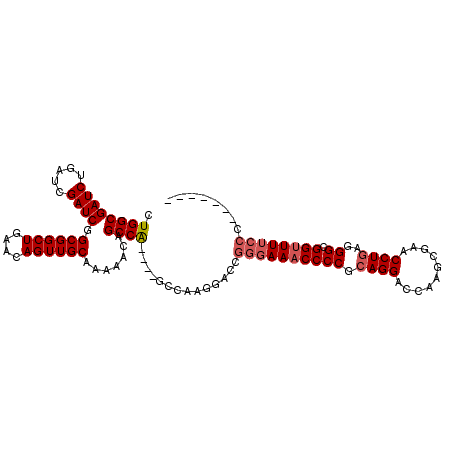

| Location | 15,514,063 – 15,514,162 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -30.88 |
| Energy contribution | -32.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514063 99 - 22224390 -------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGC----UGGCUGUUUUUGCAACUGUUCAGCCGCCGAUCGAUCAGAUCGCCAG -------((((((....(((.((((..(....)..)))).))).))))))(((....(((----.(((((.....((....)).))))))))((((.....))))))).. ( -40.60) >DroSec_CAF1 74525 99 - 1 -------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGC----UGGCUGUUUUUGCAACUGUUCAGCCGCCGAUCGAUCAGAUCGCCAG -------((((((....(((.((((..(....)..)))).))).))))))(((....(((----.(((((.....((....)).))))))))((((.....))))))).. ( -40.60) >DroSim_CAF1 86531 103 - 1 AAAA---GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGC----UGGCUGUUUUUGCAACUGUUCAGCCGCCGAUCGAUCAGAUCGCCAG ....---((((((....(((.((((..(....)..)))).))).))))))(((....(((----.(((((.....((....)).))))))))((((.....))))))).. ( -40.80) >DroEre_CAF1 76412 103 - 1 CUCAGCCGCUCAACCCGCCCUCAGGUUCGCUGGGUCCUUCGGGGUUUCCCG-------GUUCCUCGGCUGUUUUUGCAACUGUUCAGCCGCCGAUCGAUCAGAUCGCCAG ....((.(((.(((..(((...(((...(((((((((....)))...))))-------)).))).)))(((....)))...))).))).))(((((.....))))).... ( -33.30) >consensus _______GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGC____UGGCUGUUUUUGCAACUGUUCAGCCGCCGAUCGAUCAGAUCGCCAG .......((((((....(((.((((..(....)..)))).))).))))))......((((....((((((.....((....)).))))))..((((.....)))))))). (-30.88 = -32.00 + 1.12)


| Location | 15,514,103 – 15,514,199 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -36.58 |
| Energy contribution | -36.36 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514103 96 + 22224390 CAGCCAGCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC-------UUUUUCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCCA ......((..(((((.(((....)))(((((((..((.(((((.(((((.((....)))-------))))....)))))))....)))))))..))))))).. ( -37.70) >DroSec_CAF1 74565 96 + 1 CAGCCAGCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC-------UUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAU ......((..(((((.(((....)))(((((((..((.(((((..((((.((.....))-------....)))))))))))....)))))))..))))))).. ( -38.80) >DroSim_CAF1 86571 103 + 1 CAGCCAGCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCCUUUUCCCUUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAU ......((..(((((.(((....)))(((((((..((.(((((.(((((.((.....))....)))))......)))))))....)))))))..))))))).. ( -39.40) >consensus CAGCCAGCCAAGGACCGGGAAACCCCGCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC_______UUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAU ......((..(((((.(((....)))(((((((..((.(((((..((((..(...............)..)))))))))))....)))))))..))))))).. (-36.58 = -36.36 + -0.22)

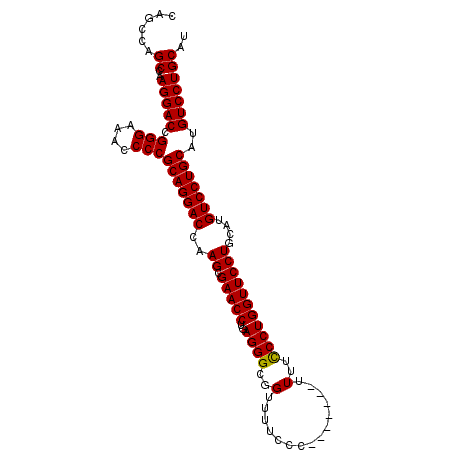

| Location | 15,514,103 – 15,514,199 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -37.87 |
| Energy contribution | -38.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514103 96 - 22224390 UGGCAGGACAUGCAGGACAUGCAGGAACCAGGAAAAA-------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGCUGGCUG .((((((((..(((((((..((..((((((((.....-------((.....))..)))..)))))))...)))))))(((....))).)))))..)))..... ( -39.60) >DroSec_CAF1 74565 96 - 1 AUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA-------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGCUGGCUG ..(((((((..(((((((..((..(((((((((....-------((.....)).))))..)))))))...)))))))(((....))).)))))..))...... ( -43.10) >DroSim_CAF1 86571 103 - 1 AUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAAGGGAAAAGGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGCUGGCUG ...(((..((.(((((((..((((((.((((((((.((((....((.....)).))))....))).)))))))))))(((....))).)))))..)))).))) ( -43.80) >consensus AUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA_______GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGCGGGGUUUCCCGGUCCUUGGCUGGCUG ..(((((((..(((((((..((..(((((((((...........((.....)).))))..)))))))...)))))))(((....))).)))))..))...... (-37.87 = -38.20 + 0.33)


| Location | 15,514,129 – 15,514,220 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -29.18 |
| Energy contribution | -28.96 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514129 91 + 22224390 GCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC-------UUUUUCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCCAGUCAGGAAGUCAACGUCACUU (((((((..((.(((((.(((((.((....)))-------))))....)))))))....)))))))...(((((.....))))).............. ( -30.90) >DroSec_CAF1 74591 91 + 1 GCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC-------UUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAUGUCAGGAAGUCAACGUCACUU (((((((((.(.(((...(((((.......)))-------)))))).))).))))))...(((((((((.....)))).))))).............. ( -32.80) >DroSim_CAF1 86597 98 + 1 GCAGGACCAAGCGAACCUGAGGGCGGUUUUCCCUUUUCCCUUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAUGUCAGGAAGUCAACGUCACUU (((((((((.(.(((...(((((.((.....))....))))))))).))).))))))...(((((((((.....)))).))))).............. ( -33.60) >consensus GCAGGACCAAGCGAACCUGAGGGCGGUUUUCCC_______UUUUCCCUGGUUCCUGCAUGUCCUGCAUGUCCUGCAUGUCAGGAAGUCAACGUCACUU (((((((..((.(((((..((((..(...............)..)))))))))))....)))))))...(((((.....))))).............. (-29.18 = -28.96 + -0.22)


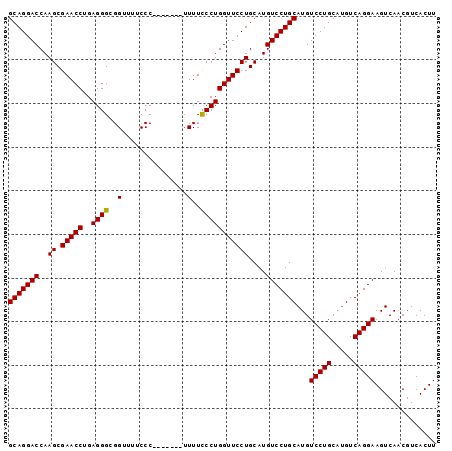
| Location | 15,514,129 – 15,514,220 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -29.37 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514129 91 - 22224390 AAGUGACGUUGACUUCCUGACUGGCAGGACAUGCAGGACAUGCAGGAACCAGGAAAAA-------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGC ..............(((((.....)))))...(((((((..((..((((((((.....-------((.....))..)))..)))))))...))))))) ( -29.80) >DroSec_CAF1 74591 91 - 1 AAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA-------GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGC ..............(((((.....)))))...(((((((..((..(((((((((....-------((.....)).))))..)))))))...))))))) ( -34.60) >DroSim_CAF1 86597 98 - 1 AAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAAGGGAAAAGGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGC ..............(((((.((((.....)))))))))...((((((.((((((((.((((....((.....)).))))....))).))))))))))) ( -37.20) >consensus AAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA_______GGGAAAACCGCCCUCAGGUUCGCUUGGUCCUGC ..............(((((.....)))))...(((((((..((..(((((((((...........((.....)).))))..)))))))...))))))) (-29.37 = -29.70 + 0.33)

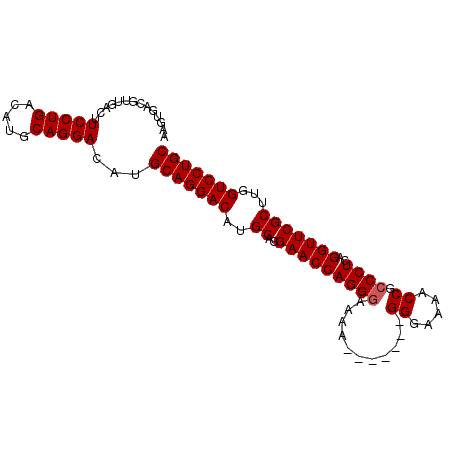

| Location | 15,514,162 – 15,514,254 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15514162 92 - 22224390 GUCUCCUGUUUUUGCACAUGUUGACACAUUCGCGAAGUGACGUUGACUUCCUGACUGGCAGGACAUGCAGGACAUGCAGGAACCAGGAAAAA--- ...(((((((((((((...((..((....((((...)))).))..)).(((((..((......))..)))))..)))))))).)))))....--- ( -28.20) >DroSec_CAF1 74624 92 - 1 GUCUCCUGUUUUUGCACAUGUUGACACAUUCGCGAAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA--- .((.((((((((((((...((..((....((((...)))).))..)).(((((.((((.....)))))))))..)))))))).))))))...--- ( -30.50) >DroSim_CAF1 86634 95 - 1 GUCUCCUGUUUUUGCACAUGUUGACACAUUCGCGAAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAAGGG .((.((((((((((((...((..((....((((...)))).))..)).(((((.((((.....)))))))))..)))))))).))))))...... ( -30.50) >consensus GUCUCCUGUUUUUGCACAUGUUGACACAUUCGCGAAGUGACGUUGACUUCCUGACAUGCAGGACAUGCAGGACAUGCAGGAACCAGGGAAAA___ ...(((((((((((((...((..((....((((...)))).))..)).(((((.((((.....)))))))))..)))))))).)))))....... (-28.09 = -28.53 + 0.45)


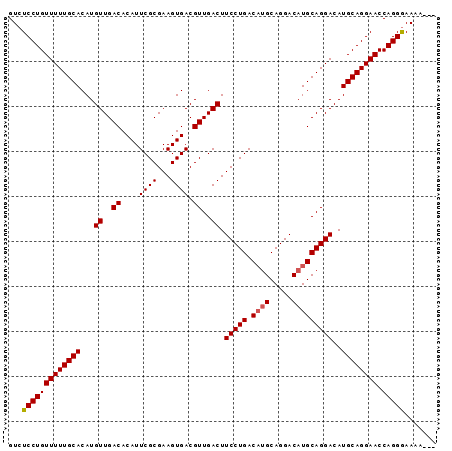
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:50 2006