
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,495,835 – 15,495,968 |
| Length | 133 |
| Max. P | 0.710224 |

| Location | 15,495,835 – 15,495,928 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -10.70 |
| Energy contribution | -12.10 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15495835 93 + 22224390 AAUCCCUGAAGUCGC-AGGCAAAACAAUUGAUCGGCGAUUUUAAGUGCACUUUUA----------------------GUGCACUUUUACUGCACUUUUAUUCGCUGCCGGC-CGGCC ......(((((((((-.(((((.....))).)).)))))))))(((((((.....----------------------))))))).....................(((...-.))). ( -25.30) >DroSec_CAF1 55939 95 + 1 AAUCCCUGUAGAAGAAAGGCAAAACAAUUGAUCGGCGAUUUUAAGUGCACUUUUA----------------------GUGCACAUUUACUGCACUUUUAUUCGCUGCCGUCCCGGCC .......(..((.((((((((....(((((.....)))))....((((((.....----------------------))))))......))).))))).))..).((((...)))). ( -22.80) >DroSim_CAF1 68564 83 + 1 AAUCCCUGUAGACGA-AGGCAAAACAAUUGAUCGGCGAUUUUAAGUGCACUUUUA----------------------GUGC-----------ACUUUUAUUCGCUGCCGUCCCGGCC ....(((........-)))..............(((((.(..((((((((.....----------------------))))-----------))))..).)))))((((...)))). ( -25.70) >DroEre_CAF1 57139 82 + 1 AAUCCCUGAAGUCGC-AGGCAAAACAAUUGAACGGCGACUUUUAGU---------------------------------GCACUUUUACUGCACUUUUAUUCGCUGC-CGCCCGGCC ..........((((.-.(((....(....)..((((((.....(((---------------------------------(((.......)))))).....)))))).-.))))))). ( -22.10) >DroYak_CAF1 62546 116 + 1 AAUCCCUGAAGUCGA-AGGCAAAACAAUUGAUCGGCGACUUUUAUUGCACUUUUACUGCACUUUUAGUGCACUUUUACUGCACUUUAACUGCACUUUUAUUCGCUGCCCGUUCGGCC ..........(((((-(((((.............((((......))))........((((.((..((((((.......))))))..)).))))...........))))..)))))). ( -26.50) >consensus AAUCCCUGAAGUCGA_AGGCAAAACAAUUGAUCGGCGAUUUUAAGUGCACUUUUA______________________GUGCACUUUUACUGCACUUUUAUUCGCUGCCGGCCCGGCC .................(((............((((((....((((((..........................................))))))....))))))........))) (-10.70 = -12.10 + 1.40)

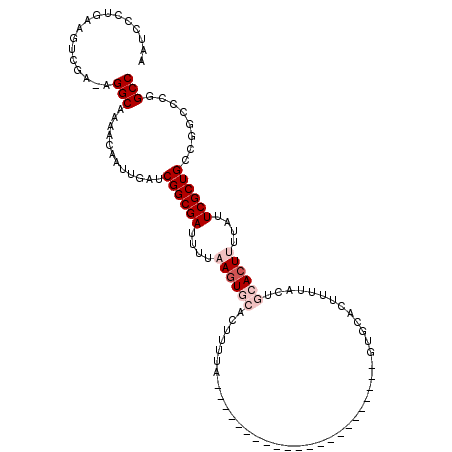

| Location | 15,495,871 – 15,495,968 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -10.95 |
| Energy contribution | -12.49 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15495871 97 + 22224390 AUUUUAAGUGCACUUUUA----------------------GUGCACUUUUACUGCACUUUUAUUCGCUGCCGGC-CGGCCCAACAAAUUUCUGGUGUUUUACGACACGGAUUUGAACUAC .....((((((((.....----------------------))))))))....................(((...-.)))....(((((((...(((((....))))))))))))...... ( -21.80) >DroSec_CAF1 55976 98 + 1 AUUUUAAGUGCACUUUUA----------------------GUGCACAUUUACUGCACUUUUAUUCGCUGCCGUCCCGGCCCAACAAAUUUCUAGUGUUUUACGACACGGAUUUGGGGUAC .......((((((.....----------------------))))))......(((.(((..((((...((((...))))..............(((((....)))))))))..)))))). ( -24.30) >DroSim_CAF1 68600 87 + 1 AUUUUAAGUGCACUUUUA----------------------GUGC-----------ACUUUUAUUCGCUGCCGUCCCGGCCCAACAAAUUUCUGGUGUUUUACGACACGGAUUUGGGGUAC .....((((((((.....----------------------))))-----------)))).(((((...((((...))))....(((((((...(((((....))))))))))))))))). ( -24.30) >DroEre_CAF1 57175 86 + 1 ACUUUUAGU---------------------------------GCACUUUUACUGCACUUUUAUUCGCUGC-CGCCCGGCCCAACAAAUUUCUGGUGUUUUACGACACGGAUUUCGGGUAC ......(((---------------------------------(((.......))))))............-.((((((.((..((......))(((((....)))))))...)))))).. ( -19.80) >DroYak_CAF1 62582 120 + 1 ACUUUUAUUGCACUUUUACUGCACUUUUAGUGCACUUUUACUGCACUUUAACUGCACUUUUAUUCGCUGCCCGUUCGGCCCAACAAAUUUCCAGUGUUUUACGACACGGAUUUGGGCUAC .........((........((((.((..((((((.......))))))..)).)))).........)).........(((((((......(((.(((((....)))))))).))))))).. ( -29.03) >consensus AUUUUAAGUGCACUUUUA______________________GUGCACUUUUACUGCACUUUUAUUCGCUGCCGGCCCGGCCCAACAAAUUUCUGGUGUUUUACGACACGGAUUUGGGGUAC .......((((((...........................))))))..........(((..((((((((......))))..............(((((....)))))))))..))).... (-10.95 = -12.49 + 1.54)


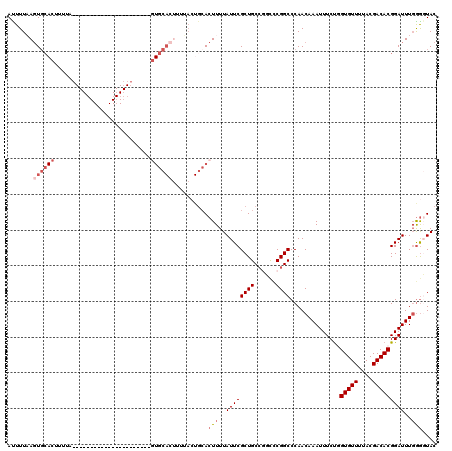
| Location | 15,495,871 – 15,495,968 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.03 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.10 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15495871 97 - 22224390 GUAGUUCAAAUCCGUGUCGUAAAACACCAGAAAUUUGUUGGGCCG-GCCGGCAGCGAAUAAAAGUGCAGUAAAAGUGCAC----------------------UAAAAGUGCACUUAAAAU (((.((.......((((......)))).....((((((...(((.-...))).))))))..)).))).....((((((((----------------------.....))))))))..... ( -24.30) >DroSec_CAF1 55976 98 - 1 GUACCCCAAAUCCGUGUCGUAAAACACUAGAAAUUUGUUGGGCCGGGACGGCAGCGAAUAAAAGUGCAGUAAAUGUGCAC----------------------UAAAAGUGCACUUAAAAU ((((.........((((......)))).....((((((...((((...)))).))))))....)))).......((((((----------------------.....))))))....... ( -24.70) >DroSim_CAF1 68600 87 - 1 GUACCCCAAAUCCGUGUCGUAAAACACCAGAAAUUUGUUGGGCCGGGACGGCAGCGAAUAAAAGU-----------GCAC----------------------UAAAAGUGCACUUAAAAU ............((((((((......((((.......))))......)))))).)).....((((-----------((((----------------------.....))))))))..... ( -23.70) >DroEre_CAF1 57175 86 - 1 GUACCCGAAAUCCGUGUCGUAAAACACCAGAAAUUUGUUGGGCCGGGCG-GCAGCGAAUAAAAGUGCAGUAAAAGUGC---------------------------------ACUAAAAGU ............((((((((......((((.......)))).....)))-))).))......((((((.......)))---------------------------------)))...... ( -18.70) >DroYak_CAF1 62582 120 - 1 GUAGCCCAAAUCCGUGUCGUAAAACACUGGAAAUUUGUUGGGCCGAACGGGCAGCGAAUAAAAGUGCAGUUAAAGUGCAGUAAAAGUGCACUAAAAGUGCAGUAAAAGUGCAAUAAAAGU ...((((...(((((((......)))).))).....(((......))))))).(((......(.((((.((..((((((.......))))))..)).)))).).....)))......... ( -31.60) >consensus GUACCCCAAAUCCGUGUCGUAAAACACCAGAAAUUUGUUGGGCCGGGCCGGCAGCGAAUAAAAGUGCAGUAAAAGUGCAC______________________UAAAAGUGCACUUAAAAU .............((((......)))).....((((((...(((.....))).))))))....((((.......)))).......................................... (-10.80 = -11.10 + 0.30)

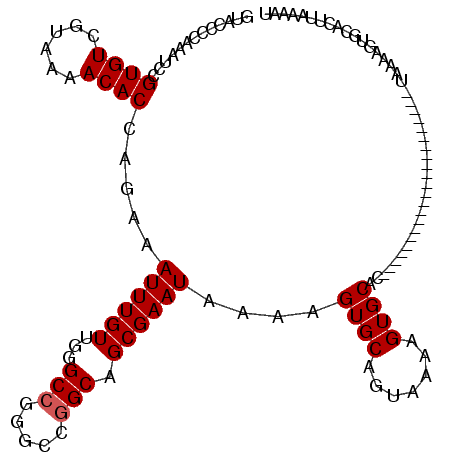
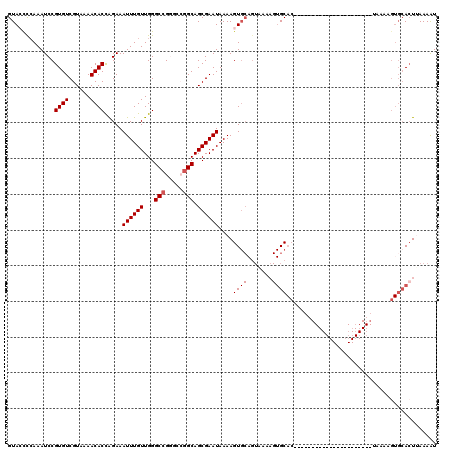
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:33 2006