
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,493,927 – 15,494,114 |
| Length | 187 |
| Max. P | 0.997206 |

| Location | 15,493,927 – 15,494,041 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -32.90 |
| Energy contribution | -33.54 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
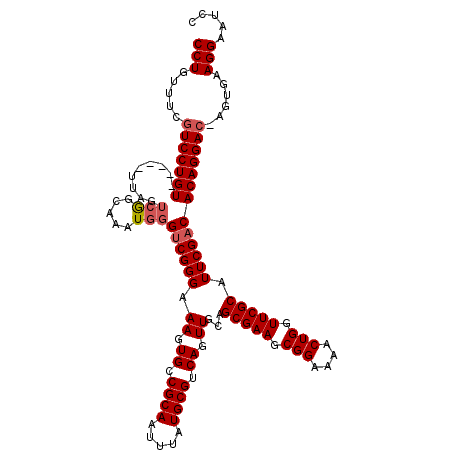
>X_DroMel_CAF1 15493927 114 + 22224390 CCUGUUUCUUCCUGU-----UUAGUCGGCAAAUGGGUCGGGAAAGUGCCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGAC-AGUGAAGGAAUCC ...((((((((((((-----((.((((((((.((((((......).)))(((.....)))..)).)))).(((((.(((....))).)))))...))))...))))-)).)))))))).. ( -38.90) >DroSec_CAF1 54070 114 + 1 CCUGUUUCGUCCUGU-----UUAGUCGGCAAAUGGGUCGGGAAAGUGCCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGAC-AGUGAAGGAAUCC (((.....(((((((-----...((((((((.((((((......).)))(((.....)))..)).)))).(((((.(((....))).)))))...)))))))))))-.....)))..... ( -39.30) >DroEre_CAF1 55322 114 + 1 CCUGUUUCGUCCUGU-----UUAGUCGGCAAAUGGGUCGGGAAAGUGGCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGAC-AGUGAAGGAAUCC (((.....(((((((-----...((((((......))).(((((.(((((((.....))))))).))...(((((.(((....))).))))).)))))))))))))-.....)))..... ( -41.00) >DroYak_CAF1 60744 114 + 1 CCUGUUUGGUCCUGU-----UUAGUCGGCAAAUGGGUCGGGAAAGUGCCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGAC-AGUGAAGGAAUCC (((.....(((((((-----...((((((((.((((((......).)))(((.....)))..)).)))).(((((.(((....))).)))))...)))))))))))-.....)))..... ( -40.40) >DroAna_CAF1 86746 120 + 1 CCUGUUUCGUCCUGUUUCGCUUAAGGAGCAAAUGGGACGGGAAAGUGCCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGACUUGCGAAGGAAUCC (((((..((((((((((.((((...))))))))))))))..)...((.((((.....)))).))((((..(((((.(((....))).)))))...)))).)))).(((....)))..... ( -41.40) >consensus CCUGUUUCGUCCUGU_____UUAGUCGGCAAAUGGGUCGGGAAAGUGCCGCAAUUUAUGCGUCAGUUGCAGCGAAGCGGAAAACUGGUUCGCAUUCGACACAGGAC_AGUGAAGGAAUCC (((.....(((((((.........(((.....)))((((((.((.((.((((.....)))).)).))...(((((.(((....))).))))).)))))))))))))......)))..... (-32.90 = -33.54 + 0.64)

| Location | 15,494,002 – 15,494,114 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15494002 112 + 22224390 AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGUCGAGUCAGGUAUGCGAAGGACUUGGAACUAAAACUGCAAAAUUAGA-------AAUGAGAGC ....((((((..(((((((((......)).....(((((((...)))))))....)))))))(((((...(....)))))))))))).....((....(((..-------..)))..)) ( -24.10) >DroSec_CAF1 54145 112 + 1 AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGUCGAGUCAGGUAUGCGAAGGACUUGGAACCAGAACUGCAAAAUUGGA-------AAUGAGAGC ...(((((((..(((((((((......)).....(((((((...)))))))....)))))))(((((...(....)))))))))))))...............-------......... ( -29.10) >DroSim_CAF1 66750 112 + 1 AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGUCGAGUCAGGUAUGCGAAGGACUUGGAACCAGAACUGCAAAAUUAGA-------AAUGAGAGC ...(((((((..(((((((((......)).....(((((((...)))))))....)))))))(((((...(....)))))))))))))....((....(((..-------..)))..)) ( -29.70) >DroEre_CAF1 55397 112 + 1 AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGCCGAGUCAGGUAUGCGGAGGACUUGGAACCAGAACUGCGAAAUUAGA-------AAUGAGAAC ...(((((((((((((..(((......)))..(((((((((...))))))).....(((((((............)))))))..)).))).)))).)))))).-------......... ( -31.60) >DroYak_CAF1 60819 119 + 1 AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGUCGAGUCAGGUAUGCGGCGGACUUGGAACCAGAACUGCGGAAUGAGAACUGAUAAAUGAGAAC .....(((((.(((((...((..(((..(((...(((((((...))))))).....))).)))..)).....((((..(((....)))..)))).))))).)))))............. ( -31.40) >consensus AAACUGGUUCGCAUUCGACACAGGACAGUGAAGGAAUCCUGCGCCAGGAUUAAAAGUCGAGUCAGGUAUGCGAAGGACUUGGAACCAGAACUGCAAAAUUAGA_______AAUGAGAGC ...(((((((..(((((((((......)).....(((((((...)))))))....)))))))(((((.........))))))))))))............................... (-26.04 = -26.28 + 0.24)


| Location | 15,494,002 – 15,494,114 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -26.84 |
| Energy contribution | -26.72 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
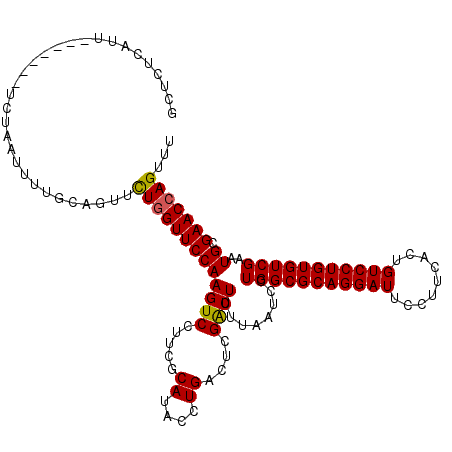
>X_DroMel_CAF1 15494002 112 - 22224390 GCUCUCAUU-------UCUAAUUUUGCAGUUUUAGUUCCAAGUCCUUCGCAUACCUGACUCGACUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU .........-------......((((((...........(((((..(((......)))...))))).......(((((((((((.........)))))))))))..))))))....... ( -23.80) >DroSec_CAF1 54145 112 - 1 GCUCUCAUU-------UCCAAUUUUGCAGUUCUGGUUCCAAGUCCUUCGCAUACCUGACUCGACUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU ((.......-------.........))....(((((((((((((..(((......)))...))))........(((((((((((.........)))))))))))..)).)))))))... ( -28.99) >DroSim_CAF1 66750 112 - 1 GCUCUCAUU-------UCUAAUUUUGCAGUUCUGGUUCCAAGUCCUUCGCAUACCUGACUCGACUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU ((.......-------.........))....(((((((((((((..(((......)))...))))........(((((((((((.........)))))))))))..)).)))))))... ( -28.99) >DroEre_CAF1 55397 112 - 1 GUUCUCAUU-------UCUAAUUUCGCAGUUCUGGUUCCAAGUCCUCCGCAUACCUGACUCGGCUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU .........-------......((((((.(((.((((..(((((.....((....))....)))))..))))..((((((((((.........)))))))))))))))))))....... ( -28.30) >DroYak_CAF1 60819 119 - 1 GUUCUCAUUUAUCAGUUCUCAUUCCGCAGUUCUGGUUCCAAGUCCGCCGCAUACCUGACUCGACUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU ..............((((.(((((((.((((..(((.....(....).....))).))))))............((((((((((.........))))))))))))))).))))...... ( -30.30) >consensus GCUCUCAUU_______UCUAAUUUUGCAGUUCUGGUUCCAAGUCCUUCGCAUACCUGACUCGACUUUUAAUCCUGGCGCAGGAUUCCUUCACUGUCCUGUGUCGAAUGCGAACCAGUUU ...............................(((((((((((((.....((....))....))))........(((((((((((.........)))))))))))..)).)))))))... (-26.84 = -26.72 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:29 2006