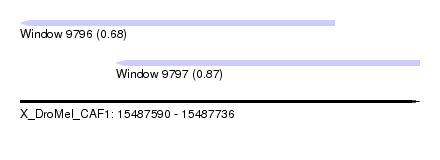
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,487,590 – 15,487,736 |
| Length | 146 |
| Max. P | 0.867933 |

| Location | 15,487,590 – 15,487,705 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.26 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15487590 115 - 22224390 UCAGUAGCGACACCUGUGGCUGUUAAACUGCACCAACUUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUUGCCGUUUGUGAAGUAAUUUGUAAGAUGCUG--UAUAUCUACA .((((((((((((((((.(((((..(((..((((......)))))))..)))....).).)))))))))))((((((...(((.....)))...)))))))))))--.......... ( -37.40) >DroSec_CAF1 47809 112 - 1 UGAAUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGCUUCGACAAACUGACCAUAGGUGUCGCUCUGCCCGUGUGUGAAGGUAUUUGUAAGAU---G--UAUAUCUACA .....(((((((((((((((.((......(((((......)))))........)).).)))))))))))))).((((..........))))..((((.(((---.--...))))))) ( -37.64) >DroSim_CAF1 60072 112 - 1 UGAAUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUGCCCGUGUGUGAAGGUACUUGUAAGAU---G--UAUAGCAAAA .....((((((((((((((.((((....)))))))...(((..((((.....))))..)))))))))))))).........(((....((((.........---)--))).)))... ( -31.50) >DroEre_CAF1 49273 115 - 1 UCAGUGGCGACAUCUAUGGCUGUAAUUAUGCACCAAGCCUGGUGGUUCUUGAAACUGACGUCAGGUGUCGCUUUUGCCGUGUGUGAACGGUCGUGUAAAAUCGUG--UAUAUAUACA .(((.((((((((((.(((.((((....)))))))((((....))))...............)))))))))).)))..(((((((.(((((........))))).--..))))))). ( -33.50) >DroYak_CAF1 48072 117 - 1 UCAGUGGCGCCAUCUAUGGCUGUAAGUCGGCACCAAGCUCGUUGGUUCUGGGAACUUACGUUAGGUGUCGCUGUUGUCGUGUGUGAGCGGUCCUGUAAAAUUAUAUCUAUAUAUAAA .(((.(.((((((.((((((....((.(((((((.....(((.(((((...))))).)))...)))))))))...)))))).))).))).).))).....((((((....)))))). ( -36.70) >consensus UCAGUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUUGCCGUGUGUGAAGGGACUUGUAAGAU__UG__UAUAUCUACA .(((.((((((((((((((.(((......))))))....((..((((.....))))..)).))))))))))).)))......................................... (-22.50 = -22.26 + -0.24)



| Location | 15,487,625 – 15,487,736 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.76 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15487625 111 - 22224390 GAGUUUAAAUGUAAA-CGCUUAAGCGAUUUGCUCAGUAGCGACACCUGUGGCUGUUAAACUGCACCAACUUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUUGCCGU ..........(((((-(((....))).))))).(((.((((((((((((.(((((..(((..((((......)))))))..)))....).).)))))))))))).))).... ( -38.20) >DroSec_CAF1 47841 111 - 1 GAGUUGAAAUGCAAA-CGCUUAAUCGAUUUGCUGAAUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGCUUCGACAAACUGACCAUAGGUGUCGCUCUGCCCGU (((((((...((...-.))....)))))))((.....(((((((((((((((.((......(((((......)))))........)).).))))))))))))))..)).... ( -40.34) >DroSim_CAF1 60104 111 - 1 GAGUUGAAAUGCAAA-CGCUUAAUCGAUUUGCUGAAUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUGCCCGU (((((((...((...-.))....)))))))((.....((((((((((((((.((((....)))))))...(((..((((.....))))..))))))))))))))..)).... ( -34.30) >DroEre_CAF1 49308 103 - 1 --UUUCAAA-------CGCUUAGGCGGCUUGCUCAGUGGCGACAUCUAUGGCUGUAAUUAUGCACCAAGCCUGGUGGUUCUUGAAACUGACGUCAGGUGUCGCUUUUGCCGU --.......-------.......(((((.........((((((((((.(((.((((....)))))))((((....))))...............))))))))))...))))) ( -29.40) >DroYak_CAF1 48109 112 - 1 GAUUUUAAAUAGAAAUAGCUGAAGUGGUGCGCUCAGUGGCGCCAUCUAUGGCUGUAAGUCGGCACCAAGCUCGUUGGUUCUGGGAACUUACGUUAGGUGUCGCUGUUGUCGU (((....(((((..(((((((.((((((((........)))))).)).)))))))....(((((((.....(((.(((((...))))).)))...))))))))))))))).. ( -37.40) >consensus GAGUUGAAAUGCAAA_CGCUUAAGCGAUUUGCUCAGUAGCGACACCUAUGGCUGUAAAACUGCACCAACCUCGGUGGUUCGACAAACUGACGAUAGGUGUCGCUCUUGCCGU ..........(((((.(((....))).))))).(((.((((((((((((((.(((......))))))....((..((((.....))))..)).))))))))))).))).... (-22.84 = -24.76 + 1.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:24 2006