
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,463,465 – 15,463,603 |
| Length | 138 |
| Max. P | 0.846296 |

| Location | 15,463,465 – 15,463,564 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.97 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.73 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15463465 99 + 22224390 GAGGCGAUGGAAAAUACACUUUGUAUGCCAGUGAAAAUGUUCGUUGGGCAUUUUCCUUUGGGUAAAAGUGUGUG------UUCUCAUUGUUUUCCGGCCAA----GUU---------G ((((((((((((.(((((((((.(((.((((.(((((((((.....)))))))))..)))))))))))))))).------))).)))))))))........----...---------. ( -33.10) >DroSec_CAF1 23561 100 + 1 -----GAUGGCAAAUACACUUUGUAUGCCAGUGGAAAUGCUCGGUGGGCAUUUUCCUUUGGGGAAAAGUGUGUG------UUCUAUUUGUUUUCCGGCCAA----GUUUAAAUA---A -----..((((..(((((((((...(.((((.(((((((((.....)))).))))).)))).).))))))))).------................)))).----.........---. ( -28.91) >DroSim_CAF1 23782 94 + 1 -----GAUGGCAAAUACACUUUGUAUGCCAGUGGAAAUGCUCGGUGGGCAUUUUCCUUUGGGGAAAAGUGUGUG------UUCUUUUUGUUUUCCGGCCAA----GUU---------G -----..((((..(((((((((...(.((((.(((((((((.....)))).))))).)))).).))))))))).------................)))).----...---------. ( -28.91) >DroYak_CAF1 24743 99 + 1 CAGGCGGUGGAAAAUACGAUUUGUACGCCAUUGGAAAUGCUCGGUGGGCAUUUUCCUUUAAGGAAAAGUGUGUG------UUGUCUUUGCUUUCCGGCCAA----GUU---------G ..(((((((...((......))...))))..((((((.((.((..(.(((((((((.....)).))))))).).------.)).....)))))))))))..----...---------. ( -27.90) >DroAna_CAF1 39175 114 + 1 GGAAAAGUGAAAAAUAUGAUUUGUGUGUCAGGAGAAAAGCUCC--GGGAAUUAUUUGU--ACAGAAAGUGUGCAAGGGAUUUUCAUUUGUUUUCCGUCCGUCCGUGAUUUAAUUCCGG (((...(.(((((...((((......))))((((.....))))--(..((((.(((((--(((.....)))))))).))))..).....)))))).)))..(((.((......))))) ( -30.10) >consensus _____GAUGGAAAAUACACUUUGUAUGCCAGUGGAAAUGCUCGGUGGGCAUUUUCCUUUGGGGAAAAGUGUGUG______UUCUAUUUGUUUUCCGGCCAA____GUU_________G .............(((((((((..........(((((((((.....))))))))).........)))))))))............................................. (-13.57 = -13.73 + 0.16)



| Location | 15,463,505 – 15,463,603 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15463505 98 - 22224390 CUUCUGGCUCCCGCUUUUCUUUUUGGACCGUUCAAAGUACAACUUGGCCGGAAAACAAUGAGAACACACACUUUUACCCAAAGGAAAAUGCCCAACGA ....(((.....(((((((((((.((.(((..(((........)))..))).......(((((........))))))).))))))))).))))).... ( -18.40) >DroSim_CAF1 23817 97 - 1 CUUCGGGCUCUCGCUUUUCUU-UUGGGCCGUUCAAAGUACAACUUGGCCGGAAAACAAAAAGAACACACACUUUUCCCCAAAGGAAAAUGCCCACCGA ....((((......(((((((-((((((((..(((........)))..)))......(((((........))))).)))))))))))).))))..... ( -28.30) >DroEre_CAF1 26476 78 - 1 C-------UCCCGAUUUUCCU-UUGGACCGUUCAAAGUACAACUUGGCCGGAAAACAAAG----CACACACUUUUCCCCAAAGGAAAGUG-------- .-------.....((((((((-((((.(((..(((........)))..))).....((((----......))))...)))))))))))).-------- ( -22.40) >DroYak_CAF1 24783 90 - 1 A-------UGCCGAUUUUCUU-UUGGACCGUUCAAAGUACAACUUGGCCGGAAAGCAAAGACAACACACACUUUUCCUUAAAGGAAAAUGCCCACCGA .-------.(((((.((..((-(((((...)))))))...)).)))))(((...(((..(......)....((((((.....)))))))))...))). ( -15.70) >consensus C_______UCCCGAUUUUCUU_UUGGACCGUUCAAAGUACAACUUGGCCGGAAAACAAAGAGAACACACACUUUUCCCCAAAGGAAAAUGCCCACCGA ..............(((((((.((((.(((..(((........)))..)))......(((((........)))))..))))))))))).......... (-12.41 = -12.47 + 0.06)

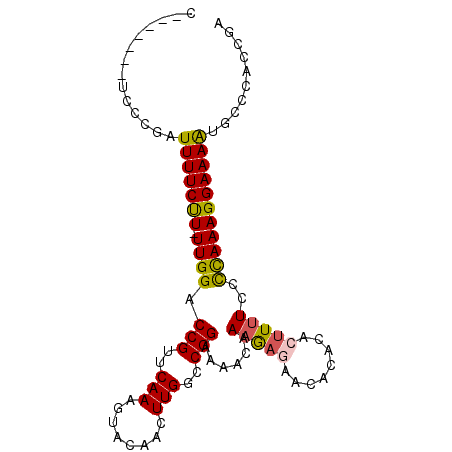
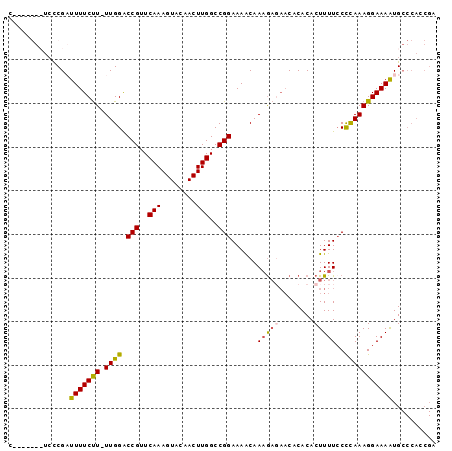
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:16 2006