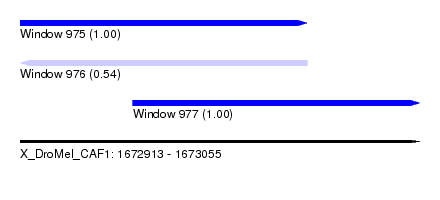
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,672,913 – 1,673,055 |
| Length | 142 |
| Max. P | 0.998533 |

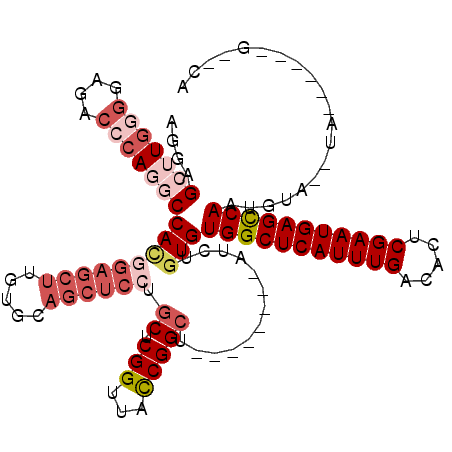
| Location | 1,672,913 – 1,673,015 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -30.88 |
| Energy contribution | -33.76 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1672913 102 + 22224390 AGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGGU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CA .(..(((((((....)))))))..)((((((.....))))))(((((((...))))))--------).((((((((((((((((.....)))))))))))...))--))------)--.. ( -52.20) >DroSec_CAF1 7021 102 + 1 AGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUGCCGGCU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CA .(..(((((((....)))))))..)((((((.....))))))....(((...)))(((--------((.....(((((((((((.....)))))))))))....)--))------)--). ( -50.90) >DroSim_CAF1 7807 102 + 1 AGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCAUGCUCGGUUGCCGGCU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CA ..(.(((((((....)))))))).(((((((.(.(((.....))).))))).)))(((--------((.....(((((((((((.....)))))))))))....)--))------)--). ( -49.00) >DroEre_CAF1 10587 106 + 1 AGGAG--UG--GAGACCCAAGCCAUGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGCU--------ACCUGUGUGGCUCAUUUGACACUCGAAUGAGUCAAUGUA--UAUUCGGUGUACA (((((--((--(.((((..(((...((((((.....)))))).))).)))).)).)))--------.)))...(((((((((((.....))))))))))).((((--(((...))))))) ( -41.10) >DroYak_CAF1 9235 105 + 1 AGCAG--UGGGGAGACGGAAGCCA-----------CAGCUCCUGCUCGGUUAUCGGCUAUCUGGCUAUCUGUGUGGCUCAUUUGACACUCGAAUGAGUCAAUGUGUAUAAUCGUGG--CA .....--...(....)....((((-----------((((....))).(((((((((((....)))).......(((((((((((.....)))))))))))....).))))))))))--). ( -34.90) >consensus AGGAGCUUGGGGAGACCCAGGCCACGGAGCUUGUGCAGCUCCUGCUCGGUUACCGGCU________AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA__UA______G__CA ....(((((((....)))))))(((((((((.....)))))).((.(((...))))).............)))(((((((((((.....))))))))))).................... (-30.88 = -33.76 + 2.88)

| Location | 1,672,913 – 1,673,015 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -20.58 |
| Energy contribution | -24.18 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1672913 102 - 22224390 UG--C------UA--UACAUUGGCUCAUUCGAGUGUCAAAUGAGCCACACAGAU--------ACCCGGUAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCU .(--(------(.--.....(((((((((.(.....).)))))))))....((.--------((((((...(((....((((((.....)))))).))).)))))).))....))).... ( -38.90) >DroSec_CAF1 7021 102 - 1 UG--C------UA--UACAUUGGCUCAUUCGAGUGUCAAAUGAGCCACACAGAU--------AGCCGGCAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCU .(--(------((--(....(((((((((.(.....).))))))))).....))--------))).(....).((((.((((((.....))))))......((((....)))).)))).. ( -40.50) >DroSim_CAF1 7807 102 - 1 UG--C------UA--UACAUUGGCUCAUUCGAGUGUCAAAUGAGCCACACAGAU--------AGCCGGCAACCGAGCAUGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCU .(--(------((--(....(((((((((.(.....).))))))))).....))--------))).(....).((((..(((((.....))))).......((((....)))).)))).. ( -36.10) >DroEre_CAF1 10587 106 - 1 UGUACACCGAAUA--UACAUUGACUCAUUCGAGUGUCAAAUGAGCCACACAGGU--------AGCCGGUAACCGAGCAGGAGCUGCACAAGCUCCAUGGCUUGGGUCUC--CA--CUCCU .....((((..((--(....((.((((((.(.....).)))))).)).....))--------)..))))..((((((.((((((.....))))))...)))))).....--..--..... ( -33.80) >DroYak_CAF1 9235 105 - 1 UG--CCACGAUUAUACACAUUGACUCAUUCGAGUGUCAAAUGAGCCACACAGAUAGCCAGAUAGCCGAUAACCGAGCAGGAGCUG-----------UGGCUUCCGUCUCCCCA--CUGCU ..--..(((..........(((((.(......).)))))..(((((((((.....((......))......((.....)).).))-----------)))))).))).......--..... ( -19.40) >consensus UG__C______UA__UACAUUGGCUCAUUCGAGUGUCAAAUGAGCCACACAGAU________AGCCGGUAACCGAGCAGGAGCUGCACAAGCUCCGUGGCCUGGGUCUCCCCAAGCUCCU ....................(((((((((.(.....).)))))))))(((.............((((.....)).)).((((((.....)))))))))((.((((....)))).)).... (-20.58 = -24.18 + 3.60)

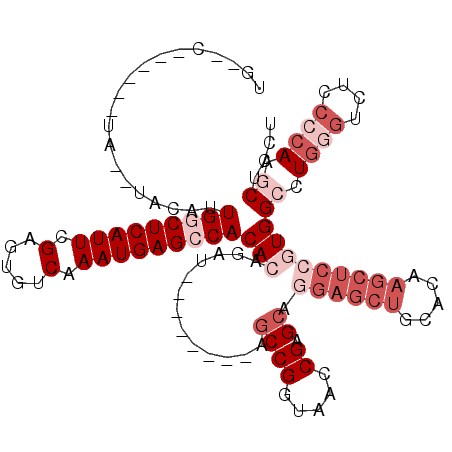
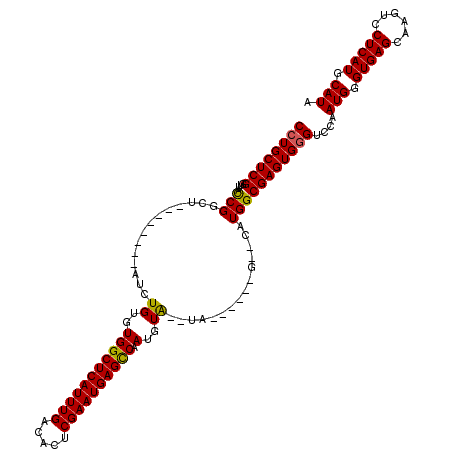
| Location | 1,672,953 – 1,673,055 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -42.42 |
| Consensus MFE | -33.65 |
| Energy contribution | -33.29 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1672953 102 + 22224390 CCUGCUCGGUUACCGGGU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUCCUCAUGCAUA ((..((((....((((.(--------((.....(((((((((((.....)))))))))))....)--))------.--).)))))))..))....(((.(((((......))))).))). ( -39.40) >DroSec_CAF1 7061 102 + 1 CCUGCUCGGUUGCCGGCU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUCCUCAUGCAUA ...(((((.(((((((((--------((.....(((((((((((.....)))))))))))....)--))------)--).)))))).)))))...(((.(((((......))))).))). ( -46.10) >DroSim_CAF1 7847 102 + 1 CAUGCUCGGUUGCCGGCU--------AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA--UA------G--CAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUCCUCAUGCAUA ...(((((.(((((((((--------((.....(((((((((((.....)))))))))))....)--))------)--).)))))).)))))...(((.(((((......))))).))). ( -46.10) >DroEre_CAF1 10623 110 + 1 CCUGCUCGGUUACCGGCU--------ACCUGUGUGGCUCAUUUGACACUCGAAUGAGUCAAUGUA--UAUUCGGUGUACAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUCCUCAUGCAUA ((..(((((...)).(((--------.......(((((((((((.....)))))))))))(((((--(((...))))))))))))))..))....(((.(((((......))))).))). ( -40.00) >DroYak_CAF1 9262 118 + 1 CCUGCUCGGUUAUCGGCUAUCUGGCUAUCUGUGUGGCUCAUUUGACACUCGAAUGAGUCAAUGUGUAUAAUCGUGG--CAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUUCUCAUGCAUA ((..((((.(.....(((((..(..((((....(((((((((((.....)))))))))))....).)))..)))))--)..).))))..))....(((.(((((......))))).))). ( -40.50) >consensus CCUGCUCGGUUACCGGCU________AUCUGUGUGGCUCAUUUGACACUCGAAUGAGCCAAUGUA__UA______G__CAUGGCGAGUGGGUCCAAUGGGUGAGCAAGUCCUCAUGCAUA ((((((((....(((..............((..(((((((((((.....)))))))))).)..))...............)))))))))))....(((.(((((......))))).))). (-33.65 = -33.29 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:12 2006