
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,433,881 – 15,433,997 |
| Length | 116 |
| Max. P | 0.950967 |

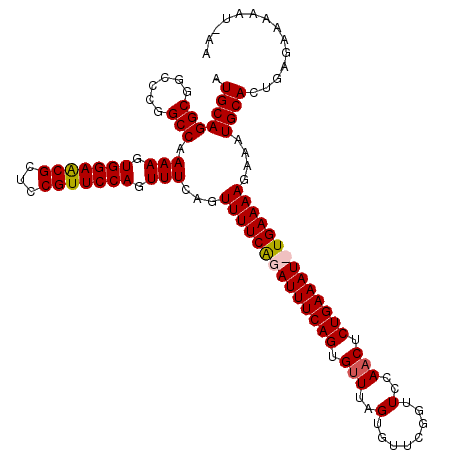
| Location | 15,433,881 – 15,433,997 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.99 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15433881 116 + 22224390 UU-AUUUUCCUCAGUGCAUUUCUUUUCA-AUUUCAGAGUUGGAACCGAACACUAAACACUGAAAUCCGAAAACUGAAACUGGAACGGAGCGCUCCACUUUUGGCCGAGCCGCCUGCAU ..-..........(((((.....(((((-.((((((..(((((.....................)))))...)))))).)))))(((..((..(((....))).))..)))..))))) ( -24.70) >DroSec_CAF1 23953 116 + 1 UC-AUUUUUCUCAGUGCACUUCUUUUCA-AUUUCAGAGUUGGAAACGAACACUAAGCACUGAAAUCCGAAAACUGAAACUGGAACGGAGCGCUCCACUUUUGGCCGAGCCGCCUGCAU ((-(.((((((((((((..(((((((((-(((....))))))))).)))......))))))).....))))).)))........(((.((((((..(....)...))).))))))... ( -28.10) >DroEre_CAF1 21822 117 + 1 UC-AUUUUGCGCAGUGCAUUUAUUUUCAAAUUUCAGCGUUGAAACCGAACACUAAACACUGAAAUCUGAAAACUAAAACUGGAACGGAGCGUUCCACUUUUGGCCGGGCCGCCUGCAU ..-.......((((.((.....((((((.(((((((.(((..............))).))))))).))))))((((((.(((((((...))))))).)))))).......)))))).. ( -35.84) >DroYak_CAF1 21908 118 + 1 UUAAUUUUUCUCAGUGCAUUUAUUUUCAAAUUUCAGAGUUGGAACUGAACACUAAACACUGAAAUCUGAAAACUGAAACUGGAACGGAGCGUUCCACUUUUGGCCGGGCCGCCUGCAU ...........(((.((.....((((((.(((((((.(((..............))).))))))).))))))(..(((.(((((((...))))))).)))..).......)))))... ( -29.64) >consensus UC_AUUUUUCUCAGUGCAUUUAUUUUCA_AUUUCAGAGUUGGAACCGAACACUAAACACUGAAAUCCGAAAACUGAAACUGGAACGGAGCGCUCCACUUUUGGCCGAGCCGCCUGCAU ...........(((.((.....((((((.(((((((.(((..............))).))))))).))))))((((((.(((((((...))))))).)))))).......)))))... (-26.36 = -26.99 + 0.63)

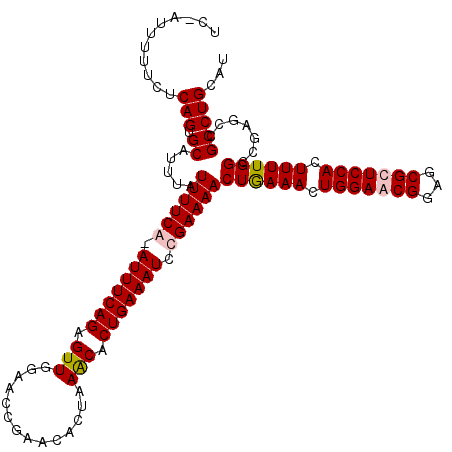

| Location | 15,433,881 – 15,433,997 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -31.15 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15433881 116 - 22224390 AUGCAGGCGGCUCGGCCAAAAGUGGAGCGCUCCGUUCCAGUUUCAGUUUUCGGAUUUCAGUGUUUAGUGUUCGGUUCCAACUCUGAAAU-UGAAAAGAAAUGCACUGAGGAAAAU-AA .....(((......))).(((.(((((((...))))))).)))..((((((....((((((((......(((((((.((....)).)))-)))).......))))))))))))))-.. ( -31.92) >DroSec_CAF1 23953 116 - 1 AUGCAGGCGGCUCGGCCAAAAGUGGAGCGCUCCGUUCCAGUUUCAGUUUUCGGAUUUCAGUGCUUAGUGUUCGUUUCCAACUCUGAAAU-UGAAAAGAAGUGCACUGAGAAAAAU-GA .....(((......))).(((.(((((((...))))))).)))...........(((((((((......((((((((.......)))).-)))).......))))))))).....-.. ( -30.52) >DroEre_CAF1 21822 117 - 1 AUGCAGGCGGCCCGGCCAAAAGUGGAACGCUCCGUUCCAGUUUUAGUUUUCAGAUUUCAGUGUUUAGUGUUCGGUUUCAACGCUGAAAUUUGAAAAUAAAUGCACUGCGCAAAAU-GA ..(((((((((...)))((((.(((((((...))))))).)))).(((((((((((((((((((.((........)).)))))))))))))))))))....)).)))).......-.. ( -45.00) >DroYak_CAF1 21908 118 - 1 AUGCAGGCGGCCCGGCCAAAAGUGGAACGCUCCGUUCCAGUUUCAGUUUUCAGAUUUCAGUGUUUAGUGUUCAGUUCCAACUCUGAAAUUUGAAAAUAAAUGCACUGAGAAAAAUUAA .(((((((......))).(((.(((((((...))))))).)))..(((((((((((((((.(((..............))).)))))))))))))))...)))).............. ( -35.14) >consensus AUGCAGGCGGCCCGGCCAAAAGUGGAACGCUCCGUUCCAGUUUCAGUUUUCAGAUUUCAGUGUUUAGUGUUCGGUUCCAACUCUGAAAU_UGAAAAGAAAUGCACUGAGAAAAAU_AA .(((((((......))).(((.(((((((...))))))).)))...((((((((((((((.(((..(........)..))).))))))))))))))....)))).............. (-31.15 = -31.40 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:08 2006