
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,399,278 – 15,399,436 |
| Length | 158 |
| Max. P | 0.873365 |

| Location | 15,399,278 – 15,399,397 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -30.23 |
| Energy contribution | -30.10 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15399278 119 + 22224390 AAGGGAGCUGACAAUUUAAUUUGCCAGCCAGGAUGUUCAAAGGACGCAGGACAACGACCUUGCCAAGUGUCCA-AGUGUAACCGAUAGGACAACAUGGAUUACAAUCCAUCCAUCCUGAA ......((((.(((......))).))))(((((((......((((((.((.(((.....)))))..)))))).-.((((..((....))...))))((((....))))...))))))).. ( -35.40) >DroSec_CAF1 45231 119 + 1 AAGGGAGCUGACAAUUUAAUUUGCCAGCCAGGAUGUUCAAAGGACGCAGGACAACGACCUUGCCAAGUGUCCA-AGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAUUCGUUCUGAA ......((((.(((......))).))))(((((((......((((((.((.(((.....)))))..)))))).-..(((((((...(((....))))).))))).......))))))).. ( -34.90) >DroEre_CAF1 36595 115 + 1 GAGGGAGCUGACAACUCAAUUUGCCAGCCAGGACGUUCAAAGGACGCAGGACAACGACCUUGCCAAGUGUUCG-AGUGUAACCAAUACGGCAACGUGGAUUACAAUCCAU----CCUGAG ......((((.(((......))).))))(((((........((((((.((.(((.....)))))..))))))(-(.(((((((...(((....))))).))))).))..)----)))).. ( -32.10) >DroYak_CAF1 39730 116 + 1 AAGGGAGCUGACAAUUUAAUUUGCCAGCCAGGACGUACAAAGGACGCAGGACAACGACCUUGCCAAGUGUCCAAAGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAU----CCUGAA ..((((((((.(((......))).))))..(((........((((((.((.(((.....)))))..))))))....(((((((...(((....))))).))))).))).)----)))... ( -35.20) >consensus AAGGGAGCUGACAAUUUAAUUUGCCAGCCAGGACGUUCAAAGGACGCAGGACAACGACCUUGCCAAGUGUCCA_AGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAU____CCUGAA .((((.((((.(((......))).))))).(((........((((((.((.(((.....)))))..))))))....(((((((...(((....))))).))))).)))......)))... (-30.23 = -30.10 + -0.13)

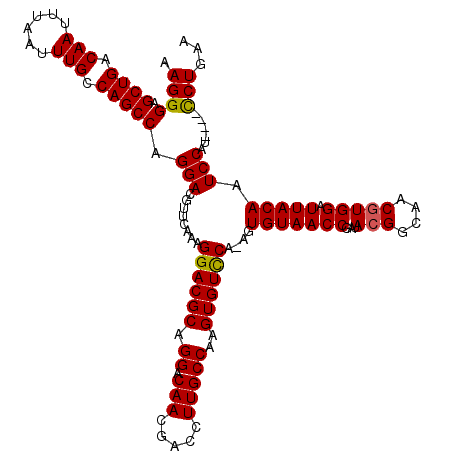

| Location | 15,399,278 – 15,399,397 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -30.95 |
| Energy contribution | -31.33 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15399278 119 - 22224390 UUCAGGAUGGAUGGAUUGUAAUCCAUGUUGUCCUAUCGGUUACACU-UGGACACUUGGCAAGGUCGUUGUCCUGCGUCCUUUGAACAUCCUGGCUGGCAAAUUAAAUUGUCAGCUCCCUU ..(((((((.((((((....))))))..(((((....(....)...-.)))))......((((.(((......))).))))....)))))))((((((((......))))))))...... ( -39.90) >DroSec_CAF1 45231 119 - 1 UUCAGAACGAAUGGAUUGUAAUCCACGUUGCCGUAUCGGUUACACU-UGGACACUUGGCAAGGUCGUUGUCCUGCGUCCUUUGAACAUCCUGGCUGGCAAAUUAAAUUGUCAGCUCCCUU ((((((.....(((((....))))).((.((((...)))).))...-.((((.(..(((((.....)))))..).))))))))))......(((((((((......)))))))))..... ( -33.60) >DroEre_CAF1 36595 115 - 1 CUCAGG----AUGGAUUGUAAUCCACGUUGCCGUAUUGGUUACACU-CGAACACUUGGCAAGGUCGUUGUCCUGCGUCCUUUGAACGUCCUGGCUGGCAAAUUGAGUUGUCAGCUCCCUC ..((((----(((((.(((((((.(((....)))...))))))).)-)...........((((.(((......))).))))....)))))))((((((((......))))))))...... ( -37.60) >DroYak_CAF1 39730 116 - 1 UUCAGG----AUGGAUUGUAAUCCACGUUGCCGUAUCGGUUACACUUUGGACACUUGGCAAGGUCGUUGUCCUGCGUCCUUUGUACGUCCUGGCUGGCAAAUUAAAUUGUCAGCUCCCUU ..((((----(((....((((((.(((....)))...))))))((...((((.(..(((((.....)))))..).))))...)).)))))))((((((((......))))))))...... ( -39.10) >consensus UUCAGG____AUGGAUUGUAAUCCACGUUGCCGUAUCGGUUACACU_UGGACACUUGGCAAGGUCGUUGUCCUGCGUCCUUUGAACAUCCUGGCUGGCAAAUUAAAUUGUCAGCUCCCUU ..((((......((((....))))(((((((((.....(((........)))...))))((((.(((......))).))))..)))))))))((((((((......))))))))...... (-30.95 = -31.33 + 0.37)

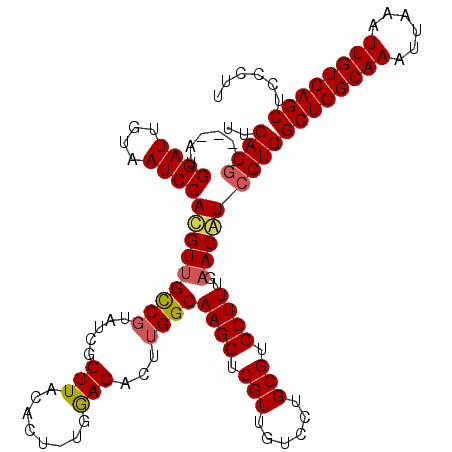
| Location | 15,399,318 – 15,399,436 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.833924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15399318 118 + 22224390 AGGACGCAGGACAACGACCUUGCCAAGUGUCCA-AGUGUAACCGAUAGGACAACAUGGAUUACAAUCCAUCCAUCCUGAAUCCAUUAGGAAUCAAAACCGAGCGAAAU-UUGCCGCACAA .((((((.((.(((.....)))))..)))))).-.((((....(((.(((.....(((((....)))))))).((((((.....)))))))))......(.((((...-))))))))).. ( -30.80) >DroSec_CAF1 45271 119 + 1 AGGACGCAGGACAACGACCUUGCCAAGUGUCCA-AGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAUUCGUUCUGAAUCCAUUAGGAAUCAAGACCGAGCGAAAUUUUGCCACACAA .((((((.((.(((.....)))))..)))))).-.((((.....))))(((((..(((((....)))))((((((((((.(((....))).))).....)))))))...)))))...... ( -36.20) >DroEre_CAF1 36635 114 + 1 AGGACGCAGGACAACGACCUUGCCAAGUGUUCG-AGUGUAACCAAUACGGCAACGUGGAUUACAAUCCAU----CCUGAGUCCAUUAGAAAUCAAAACCAACCGAAAU-UUGCCGCACAA .((((.(((((.....((.(((((..(((((.(-........))))))))))).))((((....)))).)----)))).)))).........................-........... ( -27.00) >DroYak_CAF1 39770 115 + 1 AGGACGCAGGACAACGACCUUGCCAAGUGUCCAAAGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAU----CCUGAAUCCAUUAGGAAUCAAAACCAACCGAAAU-UUACCAGACAA .(((..(((((.....((.(((((..(((((............)))))))))).))((((....)))).)----))))..)))....((........)).........-........... ( -26.40) >consensus AGGACGCAGGACAACGACCUUGCCAAGUGUCCA_AGUGUAACCGAUACGGCAACGUGGAUUACAAUCCAU____CCUGAAUCCAUUAGGAAUCAAAACCAACCGAAAU_UUGCCACACAA .((((((.((.(((.....)))))..))))))...((((.........((....((((((....))))))....))(((.(((....))).)))....................)))).. (-23.15 = -23.15 + 0.00)

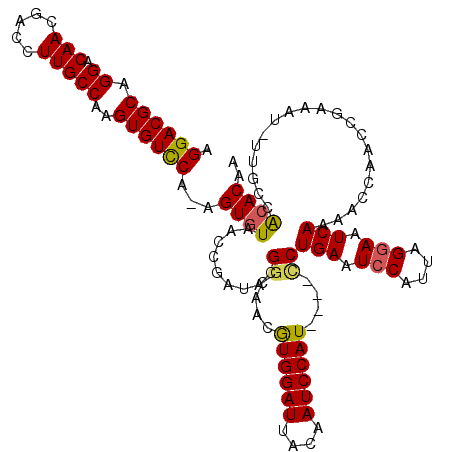
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:44 2006