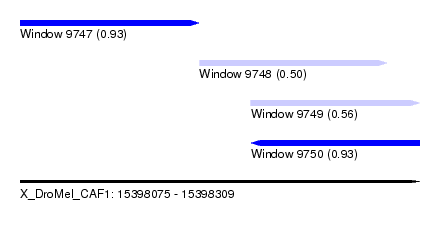
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,398,075 – 15,398,309 |
| Length | 234 |
| Max. P | 0.930754 |

| Location | 15,398,075 – 15,398,180 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15398075 105 + 22224390 CUCCACCCCCCC------CCCCU-A-CCCCCUGUUGCUUGUCAUUUGCUGCUUCAUAUUUCUUCAUUGCCAUAAUGAAGCCCAAAACAGA-------ACAGGGCAAACUGCAAUAACAAG ............------.....-.-..(((((((.((.((..((((..(((((((.................))))))).)))))))))-------))))))................. ( -19.13) >DroSec_CAF1 44084 103 + 1 ----ACUCCCAC------CCC-------CUCUGUUGCUUGUCAUUUGCUGCUUCAUAUUUAUUCAUUGCCAUAAUGAAGCCCAAAACAGAAGGCAAAACAGGGCAAACUGCAAUAACAAG ----........------.((-------((...((((((....((((..(((((((.................))))))).)))).....))))))...))))................. ( -20.13) >DroEre_CAF1 35340 113 + 1 UUCCAAUCCCCC------UUCUGGACGCCCCUGUUGCUUGUCAUUUGCUGCUUCAUAUUUCUUCAUUGCCACAAUGAAGCCCAAAACAGAAGGCA-AACAGGGCAAACUGCAAUAACAGG .((((.......------...)))).((((.((((((((....((((..(((((((.................))))))).)))).....)))).-))))))))...(((......))). ( -27.53) >DroYak_CAF1 26902 120 + 1 UUCCAAAACCCCCCUGUUUUUUGGACGCCCCUGUUGCUUGUCAUUUGCUGCUUCAUAUUUCUUCAUUGCCAUAAUGAAGCCCAAAACAGAAGGCAAAACAGGGCGAAUUGCAAUAACAGG ............((((((..(((.((((((.((((((((....((((..(((((((.................))))))).)))).....)))))..))))))))...).))).)))))) ( -31.83) >consensus UUCCAAUCCCCC______CCCUG_A_GCCCCUGUUGCUUGUCAUUUGCUGCUUCAUAUUUCUUCAUUGCCAUAAUGAAGCCCAAAACAGAAGGCA_AACAGGGCAAACUGCAAUAACAAG .............................(((((((.((((..(((((.(((((((.................)))))))((..................)))))))..))))))))))) (-19.17 = -19.30 + 0.13)

| Location | 15,398,180 – 15,398,290 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -17.85 |
| Consensus MFE | -11.51 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15398180 110 + 22224390 GGAAAAAACCAAAAAUAAAA-AUA---------AAGGAGCAAAAAAAGGACAUCCAUGCGAGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUU ((......))..........-...---------..(((.(........)...))).(((.((....((((((.((((.......((....)).....)))).))))))...))))).... ( -13.60) >DroSec_CAF1 44187 98 + 1 GGGAAAAACCAAAAAA-------A---------AAGGA--UAAAAAAGGACAUCCA-GCGGGCAAAGUAACACGUAUAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAA--- ((......))......-------.---------..(((--(..........)))).-(((((....((((((.(((........((....))......))).))))))..)))))..--- ( -19.04) >DroEre_CAF1 35453 115 + 1 GG-AAAAACCAAAGCCAAAA-AAAAGGGGGGAAAAGGA--AAAAAAUAGACAACCA-GCGGGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUU ((-.....))..........-....((..(........--..........)..)).-(((((....((((((.((((.......((....)).....)))).))))))..)))))..... ( -18.57) >DroYak_CAF1 27022 116 + 1 AG-AAAAACGAAAAAAAAAAUGAAAAAGGGGGAAAGCA--AAAAAAAGGACAACCA-UUGGGCGAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUU ..-.................((....((((((..((((--((.....((....)).-....((((..(((........))).))))..............)))))).)))))).)).... ( -20.20) >consensus GG_AAAAACCAAAAAAAAAA_AAA_________AAGGA__AAAAAAAGGACAACCA_GCGGGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUU .........................................................(((((....((((((.((((.......((....)).....)))).))))))..)))))..... (-11.51 = -12.07 + 0.56)


| Location | 15,398,210 – 15,398,309 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15398210 99 + 22224390 AAAAAAAGGACAUCCAUGCGAGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUUACAAGGGAUUUCCCCGGGG .......((....))...........((((((.((((.......((....)).....)))).))))))((((((.......)).(((.....))))))) ( -18.80) >DroSec_CAF1 44209 93 + 1 UAAAAAAGGACAUCCA-GCGGGCAAAGUAACACGUAUAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAA-----GGGGGAUUUCCCCGGGG ............(((.-(((((....((((((.(((........((....))......))).))))))..)))))..-----.((((....))))))). ( -25.44) >DroEre_CAF1 35489 98 + 1 AAAAAAUAGACAACCA-GCGGGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUUACAAGGGAUUUCCCGGGGG ..........(..((.-(((((....((((((.((((.......((....)).....)))).))))))..))))).........(((....)))))..) ( -23.80) >DroYak_CAF1 27059 98 + 1 AAAAAAAGGACAACCA-UUGGGCGAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUUACAAGGGAUUUCCCAGGGG .............((.-(((((.((.((((((.((((.......((....)).....)))).)))))).((((..........)))).)).))))))). ( -21.80) >consensus AAAAAAAGGACAACCA_GCGGGCAAAGUAACACGUAAAUUAAUCGCAAUUGCAAAAGUUAUUUGUUACCCCCUGCAAUUUACAAGGGAUUUCCCCGGGG .............((..(((((....((((((.((((.......((....)).....)))).))))))..))))).........(((....))).)).. (-18.21 = -18.78 + 0.56)



| Location | 15,398,210 – 15,398,309 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15398210 99 - 22224390 CCCCGGGGAAAUCCCUUGUAAAUUGCAGGGGGUAACAAAUAACUUUUGCAAUUGCGAUUAAUUUACGUGUUACUUUGCUCGCAUGGAUGUCCUUUUUUU ....(((((.((((..(((.(((((((((((...........)))))))))))((((.((((......))))..))))..))).)))).)))))..... ( -25.70) >DroSec_CAF1 44209 93 - 1 CCCCGGGGAAAUCCCCC-----UUGCAGGGGGUAACAAAUAACUUUUGCAAUUGCGAUUAAUAUACGUGUUACUUUGCCCGC-UGGAUGUCCUUUUUUA ((.((((.(((..((((-----.....))))((((((.(((....((((....))))....)))...))))))))).)))).-.))............. ( -25.80) >DroEre_CAF1 35489 98 - 1 CCCCCGGGAAAUCCCUUGUAAAUUGCAGGGGGUAACAAAUAACUUUUGCAAUUGCGAUUAAUUUACGUGUUACUUUGCCCGC-UGGUUGUCUAUUUUUU ((..((((....(((((((.....)))))))((((((..(((...((((....)))).....)))..))))))....)))).-.))............. ( -27.70) >DroYak_CAF1 27059 98 - 1 CCCCUGGGAAAUCCCUUGUAAAUUGCAGGGGGUAACAAAUAACUUUUGCAAUUGCGAUUAAUUUACGUGUUACUUCGCCCAA-UGGUUGUCCUUUUUUU ((..((((....(((((((.....)))))))((((((..(((...((((....)))).....)))..))))))....)))).-.))............. ( -26.70) >consensus CCCCGGGGAAAUCCCUUGUAAAUUGCAGGGGGUAACAAAUAACUUUUGCAAUUGCGAUUAAUUUACGUGUUACUUUGCCCGC_UGGAUGUCCUUUUUUU .....(((....(((((((.....)))))))((((((..(((...((((....)))).....)))..))))))....)))....((....))....... (-21.99 = -22.43 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:41 2006