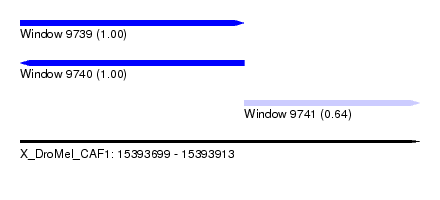
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,393,699 – 15,393,913 |
| Length | 214 |
| Max. P | 0.999595 |

| Location | 15,393,699 – 15,393,819 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -55.80 |
| Consensus MFE | -46.30 |
| Energy contribution | -48.58 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15393699 120 + 22224390 CAGUAAUUACGGGCCACGCCCCAUUCGCAUACGCCCACAUUGGCAGCUCGCAGGCAAAAGCGAUAGCGCGUGGGCGUGGCGCAGUGGGCGUGGCUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.((((((((((.((((((.(((((((((.......(((....)))....(((....))))))))))))))).))))))))))))).)))(((((((....)))))))... ( -60.10) >DroSec_CAF1 40042 120 + 1 CAGUAAUUACGGGCCACGCCCCUUUCGCAUACGCCCACAUUGACAGCUCGCAGGCAAAAGCGAUAUCGCGUGGGCGUGGCGCAGUGGGCGUGGCUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.((((((((((((..(((.(((((((((.......(((....)))....(((....))))))))))))))).)).)))))))))).)))(((((((....)))))))... ( -60.60) >DroSim_CAF1 34468 120 + 1 CAGUAAUUACGGGCCACGCCCCUUUCGCAUACGCCCACAUUGACAGCUCGCAGGCAAAAGCGAUAUCGCGUGGGCGUGGCGCAGUGGGCGUGGCUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.((((((((((((..(((.(((((((((.......(((....)))....(((....))))))))))))))).)).)))))))))).)))(((((((....)))))))... ( -60.60) >DroEre_CAF1 30921 105 + 1 CAGUAAUUACGAACCAC---------------GCCCACAUGGGCAGCUCGCAGGCAACAGCGAUAUCCUGUGGGCGGGGCGCAGUGGGCGUGGCUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.(.((((---------------((((((.((.((.(((((((((............)))))))))...)).)))))))))))).).)))(((((((....)))))))... ( -49.30) >DroYak_CAF1 22735 120 + 1 CAGUAAUUACGGGACACGCCCCUUUCGCAUACGCCCACAUUGGCAGCUCGCAGGCAAAAGCGAUAUCGUGUGGGCGGGGCGCAGUGGGCGUGGUUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.(..(((((((((..(((...(((((((((.((....((((........))))..)))))))))))..))).)).)))))))..).)))(((((((....)))))))... ( -48.40) >consensus CAGUAAUUACGGGCCACGCCCCUUUCGCAUACGCCCACAUUGGCAGCUCGCAGGCAAAAGCGAUAUCGCGUGGGCGUGGCGCAGUGGGCGUGGCUGGUAUCGUUUUGAUCAAAAUGAGUU .......(((.((((((((((....(((.(((((((((.......(((....)))....(((....)))))))))))))))....)))))))))).)))(((((((....)))))))... (-46.30 = -48.58 + 2.28)

| Location | 15,393,699 – 15,393,819 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -46.66 |
| Consensus MFE | -39.16 |
| Energy contribution | -40.80 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15393699 120 - 22224390 AACUCAUUUUGAUCAAAACGAUACCAGCCACGCCCACUGCGCCACGCCCACGCGCUAUCGCUUUUGCCUGCGAGCUGCCAAUGUGGGCGUAUGCGAAUGGGGCGUGGCCCGUAAUUACUG .....................(((..(((((((((....(((.(((((((((.((..((((........))))...))...)))))))))..)))....)))))))))..)))....... ( -52.50) >DroSec_CAF1 40042 120 - 1 AACUCAUUUUGAUCAAAACGAUACCAGCCACGCCCACUGCGCCACGCCCACGCGAUAUCGCUUUUGCCUGCGAGCUGUCAAUGUGGGCGUAUGCGAAAGGGGCGUGGCCCGUAAUUACUG .....................(((..(((((((((....(((.(((((((((.((((((((........))))..))))..)))))))))..)))....)))))))))..)))....... ( -51.60) >DroSim_CAF1 34468 120 - 1 AACUCAUUUUGAUCAAAACGAUACCAGCCACGCCCACUGCGCCACGCCCACGCGAUAUCGCUUUUGCCUGCGAGCUGUCAAUGUGGGCGUAUGCGAAAGGGGCGUGGCCCGUAAUUACUG .....................(((..(((((((((....(((.(((((((((.((((((((........))))..))))..)))))))))..)))....)))))))))..)))....... ( -51.60) >DroEre_CAF1 30921 105 - 1 AACUCAUUUUGAUCAAAACGAUACCAGCCACGCCCACUGCGCCCCGCCCACAGGAUAUCGCUGUUGCCUGCGAGCUGCCCAUGUGGGC---------------GUGGUUCGUAAUUACUG .....................(((.((((((((((((((.((...((.(.((((.((.......)))))).).)).)).)).))))))---------------)))))).)))....... ( -36.90) >DroYak_CAF1 22735 120 - 1 AACUCAUUUUGAUCAAAACGAUACCAACCACGCCCACUGCGCCCCGCCCACACGAUAUCGCUUUUGCCUGCGAGCUGCCAAUGUGGGCGUAUGCGAAAGGGGCGUGUCCCGUAAUUACUG .....................(((....(((((((....(((..((((((((.....((((........))))........))))))))...)))....)))))))....)))....... ( -40.72) >consensus AACUCAUUUUGAUCAAAACGAUACCAGCCACGCCCACUGCGCCACGCCCACGCGAUAUCGCUUUUGCCUGCGAGCUGCCAAUGUGGGCGUAUGCGAAAGGGGCGUGGCCCGUAAUUACUG .....................(((..(((((((((....(((..((((((((.....((((........))))........))))))))...)))....)))))))))..)))....... (-39.16 = -40.80 + 1.64)



| Location | 15,393,819 – 15,393,913 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -14.97 |
| Consensus MFE | -14.97 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15393819 94 + 22224390 AAAUUCCAUUUUCAUUCGCACACACGCAGCUUAGUAUGCAAAUAUGCGAUACAUUCCGUCUCGCUCACC-CUCCAUCGUGCCGUCUCUCUUCGUU .................((((....(((........)))......((((.((.....)).)))).....-.......)))).............. ( -15.00) >DroSec_CAF1 40162 95 + 1 AAAUUCCAUUUUCAUUCGCACACACGCAGCUUAGUAUGCACAUAUGCGAUACAUUCCGUCUCGCUCACCGCCCCAUAGUGCCGUCCCUCUCCAUU .................((((....(((........)))......((((.((.....)).)))).............)))).............. ( -14.90) >DroSim_CAF1 34588 95 + 1 AAAUUCCAUUUUCAUUCGCACACACGCAGCUUAGUAUGCAAAUAUGCGAUACAUUCCGUCUCGCUCACCGCCCCAUCGUGCCGUCCCUCUCCGUU .................((((....(((........)))......((((.((.....)).)))).............)))).............. ( -15.00) >consensus AAAUUCCAUUUUCAUUCGCACACACGCAGCUUAGUAUGCAAAUAUGCGAUACAUUCCGUCUCGCUCACCGCCCCAUCGUGCCGUCCCUCUCCGUU .................((((....(((........)))......((((.((.....)).)))).............)))).............. (-14.97 = -14.97 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:34 2006