
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,390,697 – 15,390,816 |
| Length | 119 |
| Max. P | 0.626561 |

| Location | 15,390,697 – 15,390,816 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15390697 119 + 22224390 AUUUUGAAAACAUAACUCGAUUCUCCACUGACCGUUGAUUUCGGUCAACGAAAUGGAAAUUCGUGCAACAAAAAGCCAAA-AAUUAUUGGCGGUCCUACCCAAAUCAACUGGCGACUGGA .......................((((.((.((((((((((.(((..(((((.(....))))))..........(((((.-.....)))))......))).)))))))).))))..)))) ( -26.20) >DroSec_CAF1 33205 119 + 1 AUUUUGGAAACAUAACUCGAUUCUCCACUGACCGUUGAUUUCGGUCCACGAAAUGGAAAUUCGUGCAACCAAAAGCCAAA-AAUUAUUGGCGGUCCUACCCGAAUCAACUGGCGACUGGA ....((....))...........((((.((.((((((((((.(((.((((((.(....))))))).........(((((.-.....)))))......))).)))))))).))))..)))) ( -31.10) >DroSim_CAF1 31330 119 + 1 AUUUUGGAAACAUAACUCGAUUCUCCACUGGCCGUUGAUUUCGGUCCACGAAAUGGAAAUUCGUGCAACAAAAAGCCAAA-AAUUAUUGGCGGUCCUACCCGAAUCAACUGGCGACUGGA ....((....))...........((((.(.(((((((((((.(((.((((((.(....))))))).........(((((.-.....)))))......))).)))))))).))).).)))) ( -36.50) >DroEre_CAF1 27712 120 + 1 AUUUUGGAAACAUAACUCGAUUCUCCACUGACCGUUGAUUACGGACCACGGAAAGGAAAUUCGUGCAACAAAAAACGAGACCAUAAUUGGCGGCCCUACCCGAAUCAACUGGCAACUGGA ....((....))...........((((.((.(((((((((.(((..((((((.......)))))).............(.(((....))))........)))))))))).))))..)))) ( -27.20) >DroYak_CAF1 19621 119 + 1 AUUUUGGAAACAUAACUCAAUUCUCCACCAACUGUUGAUUUCGGACCAAGGAAAGGAAAUUCGUGCAAUAAAAAGCCAAA-AAUUAUUGGCGGCCCUACCCGAAUCAACUGGCGACUGGA ....((....))...........(((((((...(((((((.(((.((.......))..................(((((.-.....)))))........)))))))))))))....)))) ( -24.90) >consensus AUUUUGGAAACAUAACUCGAUUCUCCACUGACCGUUGAUUUCGGUCCACGAAAUGGAAAUUCGUGCAACAAAAAGCCAAA_AAUUAUUGGCGGUCCUACCCGAAUCAACUGGCGACUGGA ....((....))...........((((....((((((((((.((..((((((.......)))))).........(((((.......))))).......)).)))))))).))....)))) (-23.26 = -23.58 + 0.32)

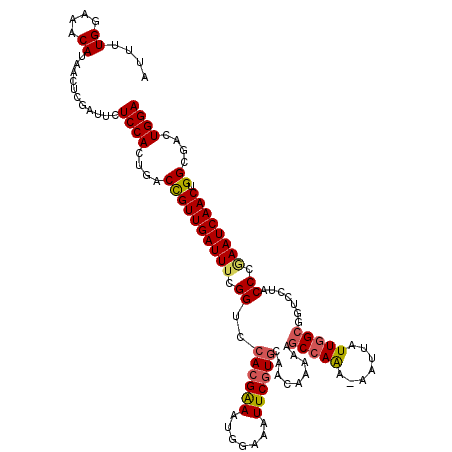

| Location | 15,390,697 – 15,390,816 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -33.72 |
| Consensus MFE | -28.18 |
| Energy contribution | -28.94 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15390697 119 - 22224390 UCCAGUCGCCAGUUGAUUUGGGUAGGACCGCCAAUAAUU-UUUGGCUUUUUGUUGCACGAAUUUCCAUUUCGUUGACCGAAAUCAACGGUCAGUGGAGAAUCGAGUUAUGUUUUCAAAAU ((((.(.(((.((((((((.(((......(((((.....-.)))))..........(((((.......)))))..))).))))))))))).).)))).....(((.......)))..... ( -34.30) >DroSec_CAF1 33205 119 - 1 UCCAGUCGCCAGUUGAUUCGGGUAGGACCGCCAAUAAUU-UUUGGCUUUUGGUUGCACGAAUUUCCAUUUCGUGGACCGAAAUCAACGGUCAGUGGAGAAUCGAGUUAUGUUUCCAAAAU ((((.(.(((.((((((((((....(((((((((.....-.)))))....)))).((((((.......))))))..))).)))))))))).).))))....................... ( -38.20) >DroSim_CAF1 31330 119 - 1 UCCAGUCGCCAGUUGAUUCGGGUAGGACCGCCAAUAAUU-UUUGGCUUUUUGUUGCACGAAUUUCCAUUUCGUGGACCGAAAUCAACGGCCAGUGGAGAAUCGAGUUAUGUUUCCAAAAU ((((.(.(((.((((((((((........(((((.....-.))))).........((((((.......))))))..))).)))))))))).).))))....................... ( -37.40) >DroEre_CAF1 27712 120 - 1 UCCAGUUGCCAGUUGAUUCGGGUAGGGCCGCCAAUUAUGGUCUCGUUUUUUGUUGCACGAAUUUCCUUUCCGUGGUCCGUAAUCAACGGUCAGUGGAGAAUCGAGUUAUGUUUCCAAAAU ((((.(.(((.(((((((((((..((((((.......))))))............((((((......)).)))).)))).)))))))))).).))))....................... ( -30.60) >DroYak_CAF1 19621 119 - 1 UCCAGUCGCCAGUUGAUUCGGGUAGGGCCGCCAAUAAUU-UUUGGCUUUUUAUUGCACGAAUUUCCUUUCCUUGGUCCGAAAUCAACAGUUGGUGGAGAAUUGAGUUAUGUUUCCAAAAU .......(((((((((((......((((((((((.....-.)))))............(((......)))...)))))..)))))))...))))(((((...........)))))..... ( -28.10) >consensus UCCAGUCGCCAGUUGAUUCGGGUAGGACCGCCAAUAAUU_UUUGGCUUUUUGUUGCACGAAUUUCCAUUUCGUGGACCGAAAUCAACGGUCAGUGGAGAAUCGAGUUAUGUUUCCAAAAU ((((.(.(((.((((((((((........(((((.......))))).........((((((.......))))))..))).)))))))))).).))))....................... (-28.18 = -28.94 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:30 2006