
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,384,775 – 15,384,932 |
| Length | 157 |
| Max. P | 0.995913 |

| Location | 15,384,775 – 15,384,892 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.581003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384775 117 - 22224390 GAAA-GUGC--AGAUGCUCAUCUGGUAUUAUCCAGAUCCAGAUGAAAUCAAAUAUUUUUUUAUUGAUUUUUUAACAAGCAAAUAGAAUCCGAGCGCGAAGCUGUUUUAAAAUGCCAAGCA ....-((((--.(((..((((((((.(((.....))))))))))).)))...(((((.(((.((((....)))).))).)))))........))))...(((..............))). ( -24.74) >DroSec_CAF1 26735 115 - 1 --AA-GUGC--AGAUGCUCAUCUGGUAUUAUACAGAUCCAGAUGAAAUCAAAUAUUUUCUUAUUGAUUUAUUAACAAACAAAUAGAAUUCGAGCGUGAAGCUGUUUUAAAAUUCCAAGCA --..-.(((--.(((..((((((((.(((.....))))))))))).)))(((((.((((...((((((((((........))))))..))))....)))).)))))...........))) ( -23.30) >DroEre_CAF1 24247 109 - 1 CAAA-GUGC--AGAUGCUCAUCUGGUAUUAU------CCAGAUGAAGUCAAAUAUUU--UUAUUGAUUUAUUAACAAACAAAUAGAAUCCGAGCGCAAAGCUGUUUUGAAAUGCCUAGCA ....-((((--.(((..((((((((......------)))))))).)))......((--(((((..(((......)))..))))))).....))))...((((............)))). ( -25.30) >DroYak_CAF1 16238 112 - 1 GAUAUGUACAUAGAUGCUCAUCUGGUAUUAU------CCAGAUGAUGUGAAAUAUUU--UUAUUGAUUUAUUAACAAACAAAUAGAAUCCGAGCACAGAGCUGUUUUAAAAUGCCUAGCA ..........((((.((((((((((......------))))))..((((......((--(((((..(((......)))..)))))))......))))))))...))))............ ( -21.30) >consensus GAAA_GUGC__AGAUGCUCAUCUGGUAUUAU______CCAGAUGAAAUCAAAUAUUU__UUAUUGAUUUAUUAACAAACAAAUAGAAUCCGAGCGCAAAGCUGUUUUAAAAUGCCAAGCA ..............(((((((((((............)))))))).....................................((((((...(((.....))))))))).........))) (-16.29 = -16.10 + -0.19)

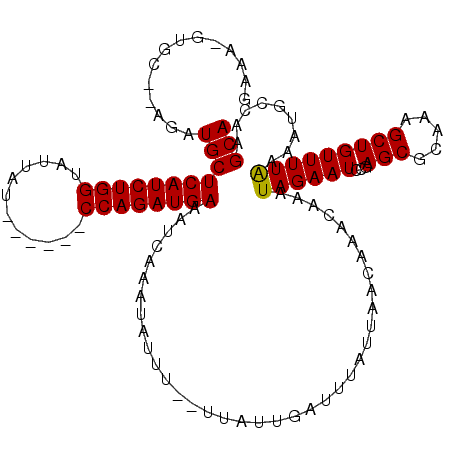
| Location | 15,384,815 – 15,384,932 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384815 117 + 22224390 UGCUUGUUAAAAAAUCAAUAAAAAAAUAUUUGAUUUCAUCUGGAUCUGGAUAAUACCAGAUGAGCAUCU--GCAC-UUUCAAUGCUAACAUUGAAUUUUGUCUAUUGGACUAGCACUUGA ((((.(((....(((..((((((........((((((((((((............))))))))).))).--....-.(((((((....)))))))))))))..))).))).))))..... ( -26.60) >DroSec_CAF1 26775 98 + 1 UGUUUGUUAAUAAAUCAAUAAGAAAAUAUUUGAUUUCAUCUGGAUCUGUAUAAUACCAGAUGAGCAUCU--GCAC-UU-------------------UUGUCUAUUCGACUAGCACUUGA ((((.(((((((...(((.(((.............((((((((............))))))))((....--)).)-))-------------------)))..)))).))).))))..... ( -17.80) >DroEre_CAF1 24287 109 + 1 UGUUUGUUAAUAAAUCAAUAA--AAAUAUUUGACUUCAUCUGG------AUAAUACCAGAUGAGCAUCU--GCAC-UUUGAAUGCCAACAUUGAAUUUUGUCUAUUGGACUAGCACUUGA ....(((((.....(((((..--...((((..(..((((((((------......))))))))((....--))..-.)..)))).....))))).....(((.....))))))))..... ( -22.40) >DroYak_CAF1 16278 112 + 1 UGUUUGUUAAUAAAUCAAUAA--AAAUAUUUCACAUCAUCUGG------AUAAUACCAGAUGAGCAUCUAUGUACAUAUCAAUGCCAGCAUUGAACUUUGUCUAUUGGACUAGCACUUGA ((((.(((....(((..((((--(.((((......((((((((------......))))))))......)))).....((((((....))))))..)))))..))).))).))))..... ( -24.00) >consensus UGUUUGUUAAUAAAUCAAUAA__AAAUAUUUGACUUCAUCUGG______AUAAUACCAGAUGAGCAUCU__GCAC_UUUCAAUGCCAACAUUGAAUUUUGUCUAUUGGACUAGCACUUGA ..............((((.................((((((((............))))))))((.............((((((....)))))).....(((.....)))..))..)))) (-15.70 = -16.32 + 0.63)

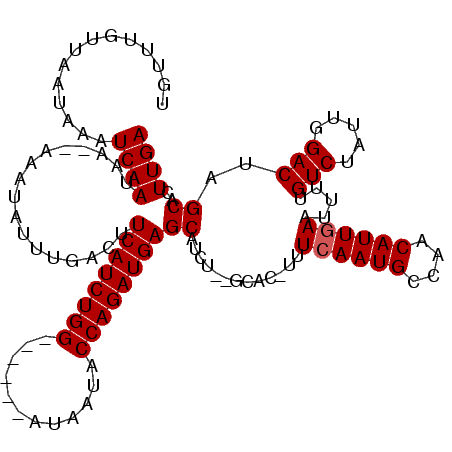

| Location | 15,384,815 – 15,384,932 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384815 117 - 22224390 UCAAGUGCUAGUCCAAUAGACAAAAUUCAAUGUUAGCAUUGAAA-GUGC--AGAUGCUCAUCUGGUAUUAUCCAGAUCCAGAUGAAAUCAAAUAUUUUUUUAUUGAUUUUUUAACAAGCA (((((..((.(((.....)))....(((((((....))))))))-)..)--.(((..((((((((.(((.....))))))))))).))).............)))).............. ( -29.60) >DroSec_CAF1 26775 98 - 1 UCAAGUGCUAGUCGAAUAGACAA-------------------AA-GUGC--AGAUGCUCAUCUGGUAUUAUACAGAUCCAGAUGAAAUCAAAUAUUUUCUUAUUGAUUUAUUAACAAACA (((((..((.(((.....)))..-------------------.)-)..)--.(((..((((((((.(((.....))))))))))).))).............)))).............. ( -22.90) >DroEre_CAF1 24287 109 - 1 UCAAGUGCUAGUCCAAUAGACAAAAUUCAAUGUUGGCAUUCAAA-GUGC--AGAUGCUCAUCUGGUAUUAU------CCAGAUGAAGUCAAAUAUUU--UUAUUGAUUUAUUAACAAACA (((((..(((((((((((............)))))).)))...)-)..)--.(((..((((((((......------)))))))).)))........--...)))).............. ( -25.10) >DroYak_CAF1 16278 112 - 1 UCAAGUGCUAGUCCAAUAGACAAAGUUCAAUGCUGGCAUUGAUAUGUACAUAGAUGCUCAUCUGGUAUUAU------CCAGAUGAUGUGAAAUAUUU--UUAUUGAUUUAUUAACAAACA ((((((((..(((.....))).....((((((....))))))...))))......((((((((((......------)))))))).)).........--...)))).............. ( -26.10) >consensus UCAAGUGCUAGUCCAAUAGACAAAAUUCAAUGUUGGCAUUGAAA_GUGC__AGAUGCUCAUCUGGUAUUAU______CCAGAUGAAAUCAAAUAUUU__UUAUUGAUUUAUUAACAAACA ((((((((..(((.....))).....((((((....))))))...))))...(((..((((((((............)))))))).))).............)))).............. (-18.36 = -18.80 + 0.44)

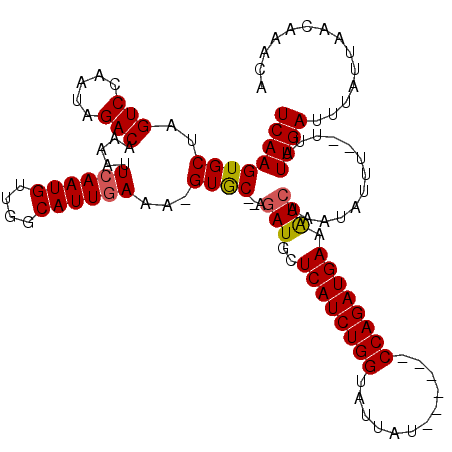
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:27 2006