
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,384,519 – 15,384,697 |
| Length | 178 |
| Max. P | 0.975765 |

| Location | 15,384,519 – 15,384,639 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -25.12 |
| Consensus MFE | -23.25 |
| Energy contribution | -24.25 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384519 120 - 22224390 AACAAGAAGGAUCGCCACAUUCUCUGGCAUAUUGUAUUUGCUUAAUGAUAUCAUAAAAGUUAAUACAUUUGUGAUUUAUAGAGAGUGAAAUAAAUGUUACCAAGCGAUUGAUAAACGAUU .........((((((..(((((((((.((((.((((((.((((.(((....)))..)))).))))))..)))).....))))))))).......((....)).))))))........... ( -27.90) >DroSec_CAF1 26478 120 - 1 AACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUGCUUAAUGAUAGCAUAAAAGUUAAUACAUUUGUGAUUUAUAGAGAGUGAAAUAAAUGUUACCAAGCGAUUGAUAAACGAUU .........((((((..(((((((((.((((.((((((.((((.(((....)))..)))).))))))..)))).....))))))))).......((....)).))))))........... ( -26.80) >DroEre_CAF1 23994 120 - 1 AACAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUUUAUUUACUUUAUGAUAGCAUAAAAGUUAAUACAUUUUUGAUUUAUAGAGAGUGAAAUAAAUGUUACCAAGCAAUUGAUAAACGAUU ((((........(((((.......)))))(((((((((..((((((((...((.(((.((....)).))).))..)))))))))))))))))..))))..(((....))).......... ( -20.20) >DroYak_CAF1 15963 120 - 1 AAUAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUGUAUUUACUUUAUGAUAGCAUAAAAGUUAAUACAUUUUUGAUUUAUAGAGAGUGAAAUAAAUGUUACCAAGCGAUUGAUAAACGAUU .........((((((..(((((((((.((...((((((.((((((((....))).))))).))))))....)).....))))))))).......((....)).))))))........... ( -25.60) >consensus AACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUACUUAAUGAUAGCAUAAAAGUUAAUACAUUUGUGAUUUAUAGAGAGUGAAAUAAAUGUUACCAAGCGAUUGAUAAACGAUU .........((((((..(((((((((.((((.((((((.((((((((....))).))))).))))))..)))).....))))))))).......((....)).))))))........... (-23.25 = -24.25 + 1.00)

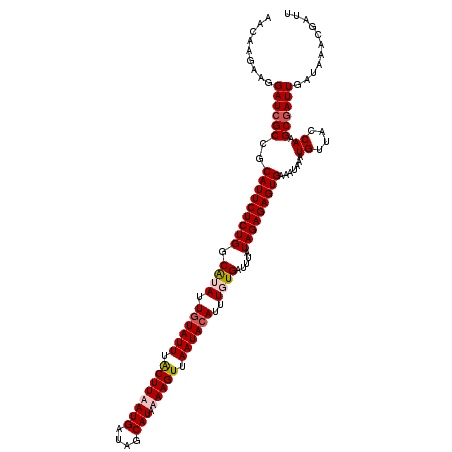

| Location | 15,384,559 – 15,384,671 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -21.20 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384559 112 + 22224390 UAUAAAUCACAAAUGUAUUAACUUUUAUGAUAUCAUUAAGCAAAUACAAUAUGCCAGAGAAUGUGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGC-------- .............((((((..(((..(((....))).)))..))))))....((((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))))-------- ( -25.10) >DroSec_CAF1 26518 112 + 1 UAUAAAUCACAAAUGUAUUAACUUUUAUGCUAUCAUUAAGCAAAUACAAUAUGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGC-------- .............((((((..(((..(((....))).)))..))))))....((((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))))-------- ( -25.40) >DroEre_CAF1 24034 112 + 1 UAUAAAUCAAAAAUGUAUUAACUUUUAUGCUAUCAUAAAGUAAAUAAAAUACGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGA-------- ...............((((.(((((.(((....)))))))).)))).......(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))).-------- ( -22.40) >DroYak_CAF1 16003 120 + 1 UAUAAAUCAAAAAUGUAUUAACUUUUAUGCUAUCAUAAAGUAAAUACAAUACGCCAGAGAAUGCGGCGAUCCUUCUUAUUGAAAAGAAAUGAUAAGCUGGCAUAUCAAUGGAGAGAAAGA ......((.....((((((.(((((.(((....)))))))).))))))...(.(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))).).)).... ( -23.40) >consensus UAUAAAUCAAAAAUGUAUUAACUUUUAUGCUAUCAUAAAGCAAAUACAAUACGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGA________ .............((((((.(((((.(((....)))))))).)))))).....(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..)))......... (-21.20 = -22.07 + 0.87)



| Location | 15,384,559 – 15,384,671 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384559 112 - 22224390 --------GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCACAUUCUCUGGCAUAUUGUAUUUGCUUAAUGAUAUCAUAAAAGUUAAUACAUUUGUGAUUUAUA --------((...((((((((((((..(((.((((((......))))))))).))).........))))))))).....))...((((.((((((((.((....)).)))))))))))). ( -26.40) >DroSec_CAF1 26518 112 - 1 --------GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUGCUUAAUGAUAGCAUAAAAGUUAAUACAUUUGUGAUUUAUA --------((((..((.((((.(((..(((.((((((......))))))))).)))))))))..))))....((((((.((((.(((....)))..)))).))))))............. ( -27.20) >DroEre_CAF1 24034 112 - 1 --------UCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUUUAUUUACUUUAUGAUAGCAUAAAAGUUAAUACAUUUUUGAUUUAUA --------.(((..((.((((.(((..(((.((((((......))))))))).)))))))))..))).(((((......((((((((....))).))))).))))).............. ( -21.20) >DroYak_CAF1 16003 120 - 1 UCUUUCUCUCCAUUGAUAUGCCAGCUUAUCAUUUCUUUUCAAUAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUGUAUUUACUUUAUGAUAGCAUAAAAGUUAAUACAUUUUUGAUUUAUA ..............((((((((((........(((((......)))))((....)).......))))))))))(((((.((((((((....))).))))).))))).............. ( -24.70) >consensus ________GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUACUUAAUGAUAGCAUAAAAGUUAAUACAUUUGUGAUUUAUA ...........(((((((((((((........(((((......)))))((....)).......))))))))))(((((.((((((((....))).))))).)))))......)))..... (-22.00 = -21.88 + -0.12)

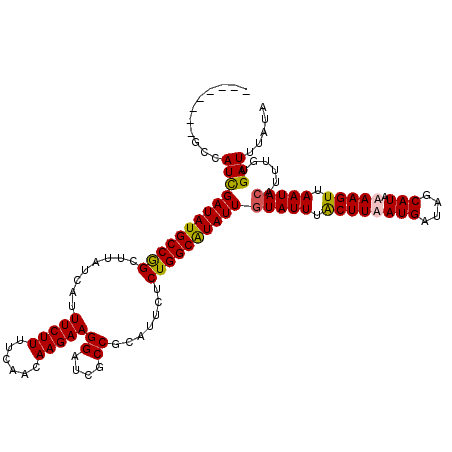
| Location | 15,384,599 – 15,384,697 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384599 98 + 22224390 CAAAUACAAUAUGCCAGAGAAUGUGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGC----------------------GAGGGAAUCCAUAAAUAAAAGAUUGA ......((((.(((((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))))----------------------).((....))...........)))). ( -23.30) >DroSec_CAF1 26558 98 + 1 CAAAUACAAUAUGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGC----------------------GCGGGAAUCCAUAAAUAAAAGAUUGA ......((((..((((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))))----------------------..((....))...........)))). ( -25.00) >DroEre_CAF1 24074 98 + 1 UAAAUAAAAUACGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGA----------------------GAGGGAAUCCAUAAAUAAAAGAUUGA ...........(.(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))).----------------------).((....))................ ( -19.80) >DroYak_CAF1 16043 120 + 1 UAAAUACAAUACGCCAGAGAAUGCGGCGAUCCUUCUUAUUGAAAAGAAAUGAUAAGCUGGCAUAUCAAUGGAGAGAAAGAGUGAGAGAUGGAGAGAGGGAAUCCAUAAAGAAAAGAUUGA ......((((.(.(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))).)..............(((((.........))))).........)))). ( -20.80) >consensus CAAAUACAAUACGCCAGAGAAUGCGGCGAUCCUUCUUGUUGAAAAGAAAUGAUAAGCCGGCAUAUCGAUGGA______________________GAGGGAAUCCAUAAAUAAAAGAUUGA ......((((...(((..(((((((((.(((.(((((......)))))..)))..))).)))).))..))).........................((....))...........)))). (-18.35 = -18.23 + -0.13)

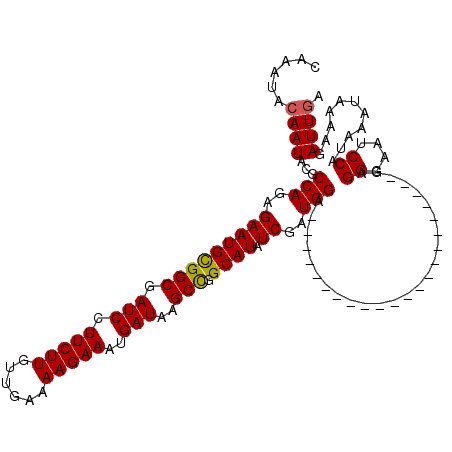
| Location | 15,384,599 – 15,384,697 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.27 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384599 98 - 22224390 UCAAUCUUUUAUUUAUGGAUUCCCUC----------------------GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCACAUUCUCUGGCAUAUUGUAUUUG ..............((((........----------------------.))))((((((((((((..(((.((((((......))))))))).))).........)))))))))...... ( -19.20) >DroSec_CAF1 26558 98 - 1 UCAAUCUUUUAUUUAUGGAUUCCCGC----------------------GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUG ................((....))..----------------------((((..((.((((.(((..(((.((((((......))))))))).)))))))))..))))............ ( -21.70) >DroEre_CAF1 24074 98 - 1 UCAAUCUUUUAUUUAUGGAUUCCCUC----------------------UCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUUUAUUUA ..............(((((.......----------------------))))).(((((((((((..(((.((((((......))))))))).)))(......).))))))))....... ( -19.00) >DroYak_CAF1 16043 120 - 1 UCAAUCUUUUCUUUAUGGAUUCCCUCUCUCCAUCUCUCACUCUUUCUCUCCAUUGAUAUGCCAGCUUAUCAUUUCUUUUCAAUAAGAAGGAUCGCCGCAUUCUCUGGCGUAUUGUAUUUA ..............(((((.........))))).....................((((((((((........(((((......)))))((....)).......))))))))))....... ( -19.40) >consensus UCAAUCUUUUAUUUAUGGAUUCCCUC______________________GCCAUCGAUAUGCCGGCUUAUCAUUUCUUUUCAACAAGAAGGAUCGCCGCAUUCUCUGGCAUAUUGUAUUUA ..(((((.........)))))................................(((((((((((........(((((......)))))((....)).......)))))))))))...... (-17.59 = -17.27 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:25 2006