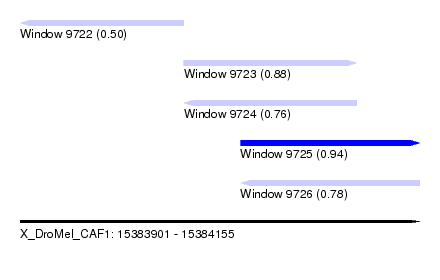
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,383,901 – 15,384,155 |
| Length | 254 |
| Max. P | 0.937061 |

| Location | 15,383,901 – 15,384,005 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15383901 104 - 22224390 CACUUUAUUACCGCAGGACAUUUGCUG------CUGCUCGUGCGUUGAGGUCCUUGCAGCAAACAAAAACAACAACUGGCAAGGAAAAACGCAAAAGGCCACAAACAACA ............((((........)))------).(((..((((((....(((((((((................)).)))))))..))))))...)))........... ( -26.09) >DroSec_CAF1 25866 110 - 1 CACUUUAUUACCGCAGGACAUUUGCUGCUGCUGCUGCUCGUGCGUUGAGGUCCUUGCCGCAAACAAAAACAACAACUGGCAAGGAAAAACGCAAAAGGCCGCAAACAACA ............((((........))))(((.(((.....((((((....(((((((((.................)))))))))..))))))...))).)))....... ( -34.23) >DroEre_CAF1 23419 104 - 1 CACUUUAUUACCGCAGGACAUUUGGUGC---UGCUGCUCGUGCGUUAAGGUCCUUGCCACAAACAAAAAC---AACAAGCAAGGAAAAAGGCAAAAUGCCACCAACAACA ............((((.((.....)).)---))).((....))(((..(((((((((.............---.....)))))))....(((.....))).))...))). ( -24.87) >consensus CACUUUAUUACCGCAGGACAUUUGCUGC___UGCUGCUCGUGCGUUGAGGUCCUUGCCGCAAACAAAAACAACAACUGGCAAGGAAAAACGCAAAAGGCCACAAACAACA ............((((.................))))...((((((....(((((((.....................)))))))..))))))................. (-18.93 = -19.26 + 0.33)


| Location | 15,384,005 – 15,384,115 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -22.66 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384005 110 + 22224390 AAAAACGAAACUCGAGGCAAGAUGGAAGCCAAAGGG------GGAACACC----CUGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGA ...........((((((((........(((.((.((------((....))----)).)).)))..(((...((((((...((....))..))))))))).......))).)))))..... ( -31.70) >DroSec_CAF1 25976 116 + 1 AAAAACGAAACUCGAGGCAAGAUGAAAGCCAAAGGAGGAGGAGGAAUACC----CCGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGA .....((((.(((..(((.........)))....))).((.((.....((----(((((((((..((....)).)))))))..)))).....)).))(((......))).))))...... ( -28.60) >DroEre_CAF1 23523 109 + 1 AAAAACGAAACUCGAGGCAAGAUGAAAGCCAGGGG-------GGAACCCC----UUUUUCGGGGAGCGUAAGCAGCGGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGA ....(((...((((((((.........)))(((((-------....))))----)...)))))...))).((((((....((....))...))))))(((......)))........... ( -36.30) >DroYak_CAF1 15460 115 + 1 AGGAACGAAACUCGAGGCAAGAUGAAAGCCAAGGGCG-----GAAACCCCCUCCCUGUUCGACGAGCAUAAGCAGCAGAAUCAGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGA .(((.((.....)).(((.........)))..(((.(-----....).))))))((((((((...((((..(((((((..((....))....)))))))......))))..)))))))). ( -33.60) >consensus AAAAACGAAACUCGAGGCAAGAUGAAAGCCAAAGG_______GGAACACC____CUGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGA .....((.....)).(((.........)))........................((((((((........(((((((...((....))..)))))))(((......)))..)))))))). (-22.66 = -22.85 + 0.19)

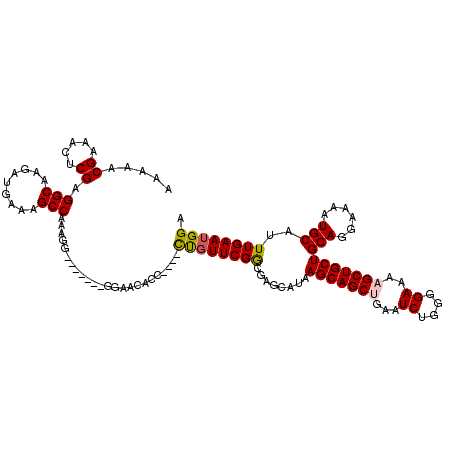

| Location | 15,384,005 – 15,384,115 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384005 110 - 22224390 UCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACAG----GGUGUUCC------CCCUUUGGCUUCCAUCUUGCCUCGAGUUUCGUUUUU .......(((((.(..(((..(((((((((..((....))...))))))........((((((..(----((....))------)..))))))........)))...)))..)))))).. ( -26.60) >DroSec_CAF1 25976 116 - 1 UCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACGG----GGUAUUCCUCCUCCUCCUUUGGCUUUCAUCUUGCCUCGAGUUUCGUUUUU .......(((((.(..(((..(((((((((..((....))...))))))........((((((.((----((..........)))).))))))........)))...)))..)))))).. ( -26.50) >DroEre_CAF1 23523 109 - 1 UCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCCGCUGCUUACGCUCCCCGAAAAA----GGGGUUCC-------CCCCUGGCUUUCAUCUUGCCUCGAGUUUCGUUUUU ..........((((......))))(((((...((....))....)))))....((((........(----((((....-------)))))(((.........)))..))))......... ( -25.40) >DroYak_CAF1 15460 115 - 1 UCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCUGAUUCUGCUGCUUAUGCUCGUCGAACAGGGAGGGGGUUUC-----CGCCCUUGGCUUUCAUCUUGCCUCGAGUUUCGUUCCU ...........((((......(((((((..((......))..)))))))..)))).....((((((((.(((....))-----).)))).(((.........))).........)))).. ( -30.70) >consensus UCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACAG____GGGGUUCC_______CCCUUGGCUUUCAUCUUGCCUCGAGUUUCGUUUUU ..........((((......))))(((((...((....))....)))))....(((((((..........))).................(((.........)))..))))......... (-18.44 = -18.25 + -0.19)



| Location | 15,384,041 – 15,384,155 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -24.13 |
| Energy contribution | -23.47 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384041 114 + 22224390 --GGAACACC----CUGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGACUACAACUCAGCCGAGGGGGUGAGGGGGCAGUCGGGGGAA --......((----((((((((........(((((((...((....))..)))))))(((......)))..)))))).((((.(..((((.((....)).))))..)..))))))))... ( -39.10) >DroSec_CAF1 26016 104 + 1 GAGGAAUACC----CCGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGACUGCAACGCAGCCGCGGGCGG------------GGGGGAA ........((----(((((((.((.((...(((((((...((....))..)))))))((((....((........))...))))......)))))))))))------------))..... ( -39.90) >DroYak_CAF1 15497 109 + 1 --GAAACCCCCUCCCUGUUCGACGAGCAUAAGCAGCAGAAUCAGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGACUGCAACUCAGCUGGGG---------GGCAGCCGGGGGAA --....((((((((.(((((...)))))...(((((((..((....))....))))))).)))..............((.((((..(((....))).---------.))))))))))).. ( -42.60) >consensus __GGAACACC____CUGUUCGGCGAGCAUAAGCAGCUGAAUCUGGGGAAAAGCUGCUGCAGGAAAAUGCAUUUGAAUGGACUGCAACUCAGCCGAGGG_G______GGCAG_CGGGGGAA ........((....((((((((........(((((((...((....))..)))))))(((......)))..)))))))).......(((....))).................))..... (-24.13 = -23.47 + -0.66)



| Location | 15,384,041 – 15,384,155 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.31 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15384041 114 - 22224390 UUCCCCCGACUGCCCCCUCACCCCCUCGGCUGAGUUGUAGUCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACAG----GGUGUUCC-- ..................(((((..(((((.((((...............((((......))))((((((..((....))...))))))....)))))))))...)----))))....-- ( -33.30) >DroSec_CAF1 26016 104 - 1 UUCCCCC------------CCGCCCGCGGCUGCGUUGCAGUCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACGG----GGUAUUCCUC .....((------------(((....((((.(((((((((...................)))))((((((..((....))...))))))..))))..))))..)))----))........ ( -33.41) >DroYak_CAF1 15497 109 - 1 UUCCCCCGGCUGCC---------CCCCAGCUGAGUUGCAGUCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCUGAUUCUGCUGCUUAUGCUCGUCGAACAGGGAGGGGGUUUC-- ..((((((((((..---------...)))))(((((((.(......)...((((......)))))))))))...(((((.(((.((.((....))..)).)))))))).)))))....-- ( -39.70) >consensus UUCCCCCG_CUGCC______C_CCCCCGGCUGAGUUGCAGUCCAUUCAAAUGCAUUUUCCUGCAGCAGCUUUUCCCCAGAUUCAGCUGCUUAUGCUCGCCGAACAG____GGUGUUCC__ ..........................((((.(((((((((...................)))))((((((..((....))...))))))....))))))))................... (-18.75 = -19.31 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:20 2006