
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,383,654 – 15,383,835 |
| Length | 181 |
| Max. P | 0.968587 |

| Location | 15,383,654 – 15,383,756 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.49 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15383654 102 - 22224390 GCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGUAGUGGCAAUGGCA---------GCACCACCA--------- .......((((((((.((((((.(((((((.(((........................))).)))).))).))))))....)))))..(((..---------...))))))--------- ( -24.46) >DroSec_CAF1 25615 104 - 1 GCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGUAGUGGCAAUGGCAAU-------GCAACAACA--------- (((.......(((((.((((((.(((((((.(((........................))).)))).))).))))))....)))))........)-------)).......--------- ( -23.32) >DroEre_CAF1 23153 120 - 1 GCAACAAUGGGCCACAAUUUGCUGCUUUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCAGCAAGUGGUAGUGGCAAUGGCAGCAGCAGCCCCACCACGACCACCAAUA .......((((((((...(((((((((....(((........)))((((((......)))))))))))))))...))))).((((....(((.......)))...)))).....)))... ( -30.90) >consensus GCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGUAGUGGCAAUGGCA_________GCACCACCA_________ ((....((..(((((.((((((.(((((((.(((........................))).)))).))).))))))....))))).)).))............................ (-21.71 = -21.49 + -0.22)

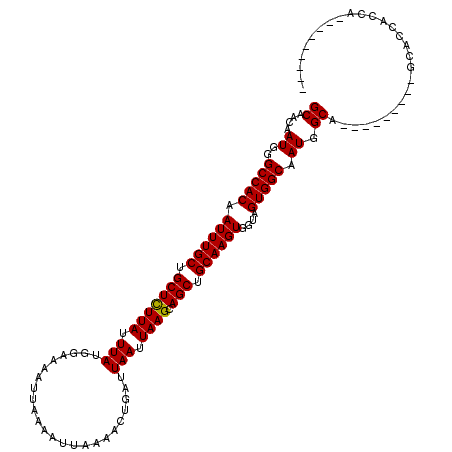
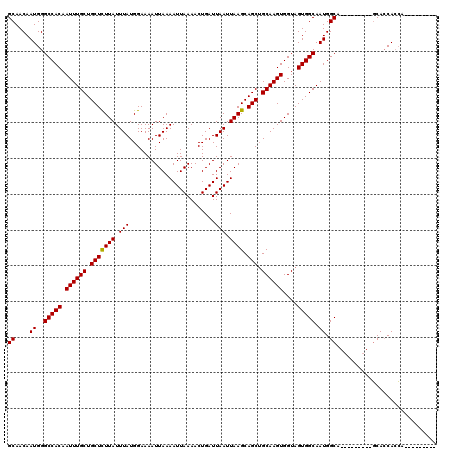
| Location | 15,383,676 – 15,383,795 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -29.92 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15383676 119 + 22224390 ACCACUUGCAGCUGCUUAAUUAAUCAGUUUUAAUUUUAAUUUUCCAUAAAUAAGAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAA-AAUGGG .(((.((((((((((((((((((......))))))...((((.....))))..)))))))...(((((((..(((((((.....)))))))))))))).........)).)))-..))). ( -30.90) >DroSec_CAF1 25639 120 + 1 ACCACUUGCAGCUGCUUAAUUAAUCAGUUUUAAUUUUAAUUUUCCAUAAAUAAGAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAAAAAUGGG .(((.((((((((((((((((((......))))))...((((.....))))..)))))))...(((((((..(((((((.....)))))))))))))).........)).)))...))). ( -30.60) >DroEre_CAF1 23193 119 + 1 ACCACUUGCUGCUGCUUAAUUAAUCAGUUUUAAUUUUAAUUUUCCAUAAAUAAAAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAGAAGUUGAAUGUCAA-AAUGGG .((((((..((((((((((((((......))))))...((((.....))))..))))))))..(((((((..(((((((.....)))))))))))))).)))(((.....)))-..))). ( -30.00) >consensus ACCACUUGCAGCUGCUUAAUUAAUCAGUUUUAAUUUUAAUUUUCCAUAAAUAAGAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAA_AAUGGG .(((.((((.(((((((((((((......))))))...((((.....))))..)))))))...(((((((..(((((((.....))))))))))))))..........).)))...))). (-29.92 = -29.70 + -0.22)

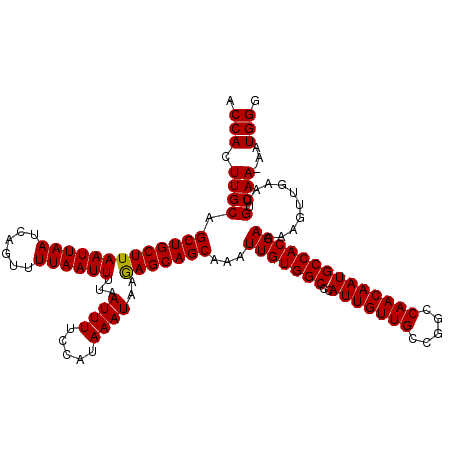
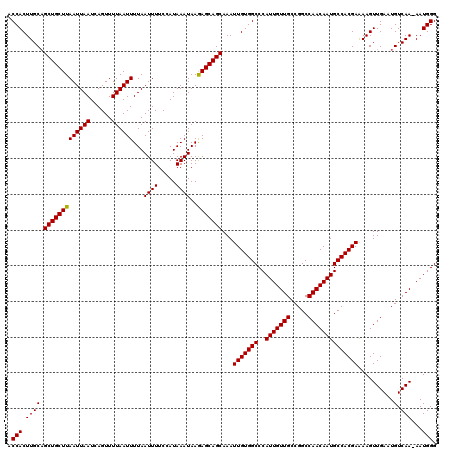
| Location | 15,383,676 – 15,383,795 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.03 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15383676 119 - 22224390 CCCAUU-UUGACAUUCAACUUUUCGUGGCAUUGUUGGCCGGCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGU .(((((-(.(.((...........((((((((((((.....)))))))..))))).....((((((((.......))).......((((((......))))))..)))))))))))))). ( -28.90) >DroSec_CAF1 25639 120 - 1 CCCAUUUUUGACAUUCAACUUUUCGUGGCAUUGUUGGCCGGCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGU .((((((.................((((((((((((.....)))))))..))))).....((((((((.......))).......((((((......))))))..)))))...)))))). ( -28.60) >DroEre_CAF1 23193 119 - 1 CCCAUU-UUGACAUUCAACUUCUCGUGGCAUUGUUGGCCGGCAACAAUGGGCCACAAUUUGCUGCUUUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCAGCAAGUGGU .(((((-(................((((((((((((.....)))))))..)))))...(((((((((....(((........)))((((((......))))))))))))))).)))))). ( -30.10) >consensus CCCAUU_UUGACAUUCAACUUUUCGUGGCAUUGUUGGCCGGCAACAAUGGGCCACAAUUUGCUGCUCUUAUUUAUGGAAAAUUAAAAUUAAAACUGAUUAAUUAAGCAGCUGCAAGUGGU .((((..(((.(............((((((((((((.....)))))))..))))).....((((((((.......))).......((((((......))))))..))))).)))))))). (-27.25 = -27.03 + -0.22)


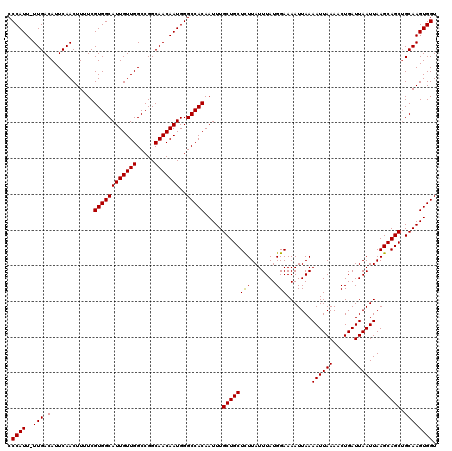
| Location | 15,383,716 – 15,383,835 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -28.43 |
| Energy contribution | -28.31 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15383716 119 + 22224390 UUUCCAUAAAUAAGAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAA-AAUGGGAGCGACCCUUCAAAGGCACAGAAAACUGCAGGGAAAAACC (((((((.........((((...(((((((..(((((((.....))))))))))))))...))))........-.)))))))...((((......(((........)))))))....... ( -29.97) >DroSec_CAF1 25679 120 + 1 UUUCCAUAAAUAAGAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAAAAAUGGGAGCGACCCUUCAAAGGCACAGAAAACUGCAGGGGAAAACC (((((((.........((((...(((((((..(((((((.....))))))))))))))...))))..........)))))))...(((((.....(((........))))))))...... ( -30.41) >DroEre_CAF1 23233 119 + 1 UUUCCAUAAAUAAAAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAGAAGUUGAAUGUCAA-AAUGGGAGCGGCCCUUCAAAGGCACAGAAAACUGCAGCGGCCAACC (((((((.........((((...(((((((..(((((((.....))))))))))))))...))))........-.))))))).((((((....))((.(((....)))..)))))).... ( -35.17) >DroYak_CAF1 15170 119 + 1 UUUCCAUAAAUAAAAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAA-AAUGGAAACGACCCUUCAAAGGCACAGAAAACUGCAGGGGAAAACC (((((((.........((((...(((((((..(((((((.....))))))))))))))...))))........-.)))))))...(((((.....(((........))))))))...... ( -31.77) >consensus UUUCCAUAAAUAAAAGCAGCAAAUUGUGGCCCAUUGUUGCCGGCCAACAAUGCCACGAAAAGUUGAAUGUCAA_AAUGGGAGCGACCCUUCAAAGGCACAGAAAACUGCAGGGGAAAACC (((((((.........((((...(((((((..(((((((.....))))))))))))))...))))..........)))))))...((((......(((........)))))))....... (-28.43 = -28.31 + -0.12)

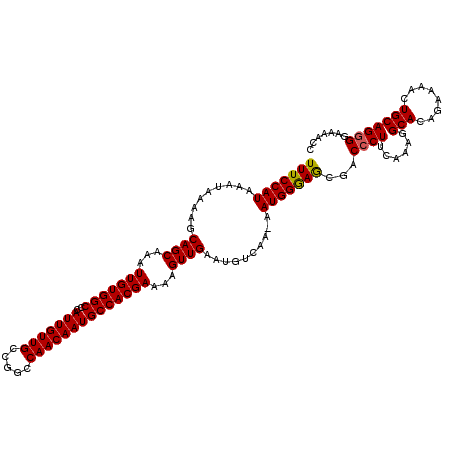

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:17 2006