
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,382,852 – 15,383,026 |
| Length | 174 |
| Max. P | 0.999992 |

| Location | 15,382,852 – 15,382,972 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.52 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.47 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.83 |
| SVM RNA-class probability | 0.999954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382852 120 + 22224390 AAUGUUUUCUUUAACAUUUCAGGUAGCAUUUGCCUUGAUCGUGAGACUCGGACUUGAAGUGAGCGUUUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAU .((((((.((((.........(((((...)))))(((((..((((((((((.((((......((......)).(((......))))))))))))))))))))))......)))))))))) ( -37.80) >DroSec_CAF1 24822 101 + 1 ------------------UCAGGUAGCAUUUGGCUUCGAAGUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAA-AACUGCGAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAU ------------------....((..(.(((((.((.((..((((((((((((((.....))))......((((.....-..))))...)))))))))).)).)).)))))..)..)).. ( -31.60) >DroEre_CAF1 22375 105 + 1 AGUGAUAU-UUUAUUAUUGCGGGGAGCAUUUGCCCUAGUCGUGAGACUCGGGCUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA--------------U .(..(((.-.....)))..).(((.((....))))).(...((((((((((((((.............)))).((((.....))))...))))))))))..)...--------------. ( -34.72) >DroYak_CAF1 14471 96 + 1 AGU----U-UUUACUUUUG------------------GUCGUGAGACUCUGAUUUGAAGUGAGUGUAUGUGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA-CCAAAAAGAGACAU .((----(-(((..(((((------------------((..((((((((..............(((....)))((((.....)))).....)))))))).....)-)))))))))))).. ( -27.30) >consensus AGU____U_UUUA_UAUUGCAGGUAGCAUUUGCCUU_GUCGUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA_CCAAAAAGAGACAU .........................................((((((((((.((((......((......)).(((......)))))))))))))))))..................... (-22.54 = -22.47 + -0.06)

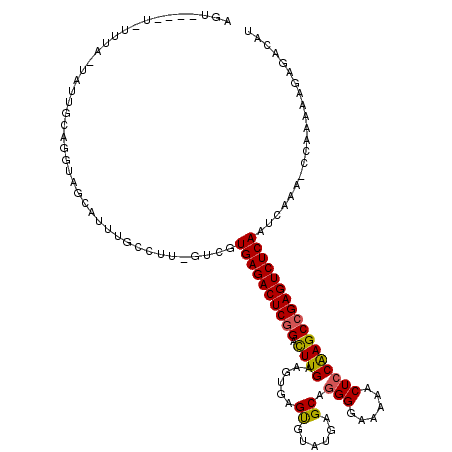

| Location | 15,382,852 – 15,382,972 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.52 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.66 |
| SVM decision value | 5.70 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382852 120 - 22224390 AUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAAACGCUCACUUCAAGUCCGAGUCUCACGAUCAAGGCAAAUGCUACCUGAAAUGUUAAAGAAAACAUU .............((((((((((((((((((((((((((.........((......))..)))))))).))))))))))..)))))))).............((((((......)))))) ( -37.10) >DroSec_CAF1 24822 101 - 1 AUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUCGCAGUU-UUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCACUUCGAAGCCAAAUGCUACCUGA------------------ .........(((((((((((.((((((((((...((((..-.....))))......(((.......)))))))))))))..)))))))))))..........------------------ ( -33.80) >DroEre_CAF1 22375 105 - 1 A--------------UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGCCCGAGUCUCACGACUAGGGCAAAUGCUCCCCGCAAUAAUAAA-AUAUCACU .--------------..((((((((((((((((((((((.....................)))))))).))))))))))......((((....))))..............-..)))).. ( -31.40) >DroYak_CAF1 14471 96 - 1 AUGUCUCUUUUUGG-UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCACAUACACUCACUUCAAAUCAGAGUCUCACGAC------------------CAAAAGUAAA-A----ACU .......(((((((-((....(((((((((..(((((((.....................)))))))..).)))))))).)))------------------))))))....-.----... ( -25.70) >consensus AUGUCUCUUUUUGG_UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCACGAC_AAGGCAAAUGCUACCUGAAAUA_UAAA_A____ACU .....................((((((((((((((((((.....................)))))))))).))))))))......................................... (-21.01 = -21.58 + 0.56)

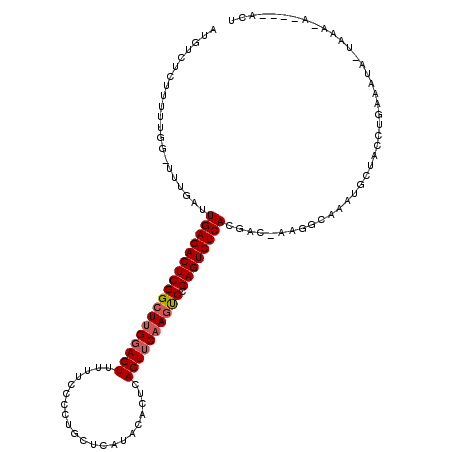

| Location | 15,382,892 – 15,382,993 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.28 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382892 101 + 22224390 GUGAGACUCGGACUUGAAGUGAGCGUUUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUC-------------------UAAGUCGGCUCAACUCUGAU .((((((((((.((((......((......)).(((......)))))))))))))))))................(((...-------------------...))).............. ( -29.70) >DroSec_CAF1 24844 119 + 1 GUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAA-AACUGCGAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUAGAGAUAAGAUGCCGAAUUAGAAUUCGGCUCAACUCUGAU .((((((((((((((.....))))......((((.....-..))))...))))))))))....................(((((....((.(((((((....)))))))))..))))).. ( -40.00) >DroEre_CAF1 22414 87 + 1 GUGAGACUCGGGCUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA--------------UC-------------------GAAUUCGGCUGAACUCUGAU .((((((((((((((.............)))).((((.....))))...)))))))))).(((..--------------((-------------------(....))).)))........ ( -27.22) >DroYak_CAF1 14488 100 + 1 GUGAGACUCUGAUUUGAAGUGAGUGUAUGUGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA-CCAAAAAGAGACAUC-------------------UAAUUCGGCUCAACUCUGAU ..(((((((...........)))).........((((.....)))).(((((((((((.......-.......)))))...-------------------....))))))...))).... ( -21.45) >consensus GUGAGACUCGGACUUGAAGUGAGUGUAUGAGCAGGGGAAAAACUCCAAGCCGAGUCUCAAUCAAA_CCAAAAAGAGACAUC___________________GAAUUCGGCUCAACUCUGAU .((((((((((.((((......((......)).(((......))))))))))))))))).............................................((((.......)))). (-23.34 = -23.28 + -0.06)

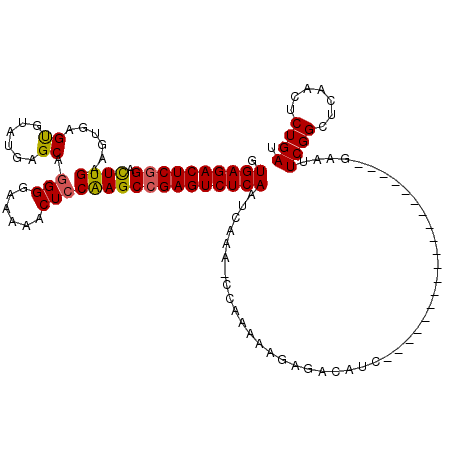
| Location | 15,382,892 – 15,382,993 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382892 101 - 22224390 AUCAGAGUUGAGCCGACUUA-------------------GAUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAAACGCUCACUUCAAGUCCGAGUCUCAC (((((((..(((..(((...-------------------...)))....)))..)))))))((((((((((((((((((.........((......))..)))))))).)))))))))). ( -35.50) >DroSec_CAF1 24844 119 - 1 AUCAGAGUUGAGCCGAAUUCUAAUUCGGCAUCUUAUCUCUAUGUCUCUUUUUGGUUUUGAUUGAGACUCGGCUCGCAGUU-UUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCAC ...((((..(((((((((....))))))).))....)))).....................((((((((((...((((..-.....))))......(((.......))))))))))))). ( -34.60) >DroEre_CAF1 22414 87 - 1 AUCAGAGUUCAGCCGAAUUC-------------------GA--------------UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGCCCGAGUCUCAC ....((((((....))))))-------------------..--------------......((((((((((((((((((.....................)))))))).)))))))))). ( -31.70) >DroYak_CAF1 14488 100 - 1 AUCAGAGUUGAGCCGAAUUA-------------------GAUGUCUCUUUUUGG-UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCACAUACACUCACUUCAAAUCAGAGUCUCAC .....(((..(((((((..(-------------------((....))).)))))-))..)))((((((((..(((((((.....................)))))))..).))))))).. ( -29.60) >consensus AUCAGAGUUGAGCCGAAUUA___________________GAUGUCUCUUUUUGG_UUUGAUUGAGACUCGGCUUGGAGUUUUUCCCCUGCUCAUACACUCACUUCAAGUCCGAGUCUCAC .............................................................((((((((((((((((((.....................)))))))))).)))))))). (-21.01 = -21.58 + 0.56)

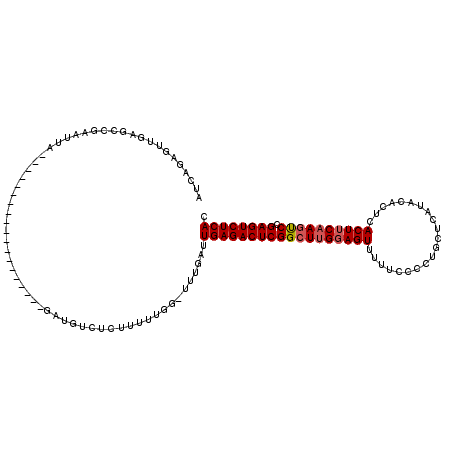
| Location | 15,382,932 – 15,383,026 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -12.76 |
| Energy contribution | -13.01 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382932 94 + 22224390 AACUCCAAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUC-------------------UAAGUCGGCUCAACUCUGAUCGGCAAUCGAGAUCGCGACGACAGCAGCAACAA------- ..(((...((((((((((...............)))))...-------------------....((((.......)))))))))....)))...((..(....)..)).....------- ( -19.66) >DroSec_CAF1 24883 113 + 1 AACUGCGAGCCGAGUCUCAAUCAAAACCAAAAAGAGACAUAGAGAUAAGAUGCCGAAUUAGAAUUCGGCUCAACUCUGAUCGGCAGUUGAGAUCGCGACGACAGCAGCAACAA------- ..((((..((((((((((...............))))).(((((....((.(((((((....)))))))))..))))).))))).((((......))))....))))......------- ( -37.56) >DroEre_CAF1 22454 87 + 1 AACUCCAAGCCGAGUCUCAAUCAAA--------------UC-------------------GAAUUCGGCUGAACUCUGAUCGGCAGUUGAGAUCGCGACGACAGCAGCAACAUCAGCAGU .......((((((((.((.......--------------..-------------------))))))))))..((((((((..((.((((...(((...))))))).))...))))).))) ( -22.10) >DroYak_CAF1 14528 100 + 1 AACUCCAAGCCGAGUCUCAAUCAAA-CCAAAAAGAGACAUC-------------------UAAUUCGGCUCAACUCUGAUCGCCAGUUGAGAUCGCGACGACAGCAGCAACAUUAGCAGU .......(((((((((((.......-.......)))))...-------------------....))))))..((((((((.(...((((...(((...)))...))))..)))))).))) ( -19.05) >consensus AACUCCAAGCCGAGUCUCAAUCAAA_CCAAAAAGAGACAUC___________________GAAUUCGGCUCAACUCUGAUCGGCAGUUGAGAUCGCGACGACAGCAGCAACAA_______ .......((((((((...............................................)))))))).......((((.........))))((..(....)..))............ (-12.76 = -13.01 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:12 2006