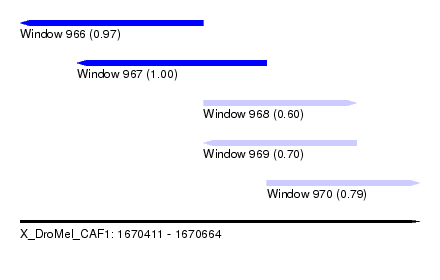
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,670,411 – 1,670,664 |
| Length | 253 |
| Max. P | 0.998696 |

| Location | 1,670,411 – 1,670,527 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.96 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1670411 116 - 22224390 UCGGUAUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGGGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUU---- ((((((..((((...(((((....))))).((((((....(((....((((....))))...)))....))))))..(((..........))).)))).))))))...........---- ( -24.30) >DroSec_CAF1 4478 116 - 1 UCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUU---- ((((((..(((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))((((.((.....)).)).)).))))))...........---- ( -25.50) >DroSim_CAF1 5330 116 - 1 UCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUU---- ((((((..(((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))((((.((.....)).)).)).))))))...........---- ( -25.50) >DroEre_CAF1 8106 120 - 1 UCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUUCAUC ((((((..(((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))((((.((.....)).)).)).))))))............... ( -25.50) >DroYak_CAF1 5508 116 - 1 UCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUU---- ((((((..(((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))((((.((.....)).)).)).))))))...........---- ( -25.50) >consensus UCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGCCUCGUUUUUUGCCAGAGCUUACUGAACCCAACCCUU____ ((((((..(((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))((((.((.....)).)).)).))))))............... (-24.28 = -24.32 + 0.04)

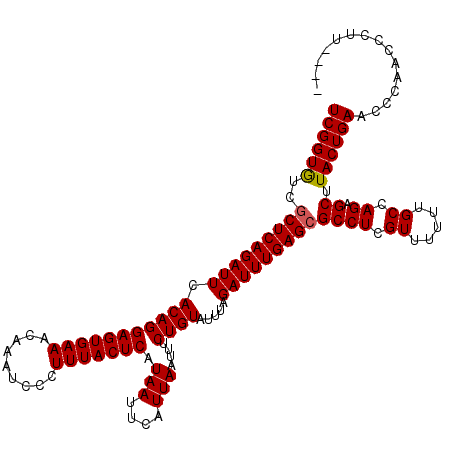
| Location | 1,670,447 – 1,670,567 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -33.82 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1670447 120 - 22224390 UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUGUCGGUAUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGGGC ....(((((((((((((((......)))..((....)).))))))))).((((((((.(((((((((((.........))))))).(((....)))....)))).....))))))))))) ( -36.60) >DroSec_CAF1 4514 120 - 1 UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCAAAAGGUGUCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGC ..(((((((((((((((((......)))))).....)))))))))))((((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))). ( -37.70) >DroSim_CAF1 5366 120 - 1 UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUGUCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGC ..(((((((((((((((((......)))))).....)))))))))))((((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))). ( -37.70) >DroEre_CAF1 8146 118 - 1 U--UGCCGAUGCCGAUGGGCUUGGGUCCGUCCGAAAGGUGUCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGC .--.((((((((((((((((....))))))).....)))))))))..((((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))). ( -40.80) >DroYak_CAF1 5544 117 - 1 U--UGCCGAUGGC-AUGGGUUUGUGUCCGUCCGUAAGGUGUCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGC .--.(((((((((-((......)))))...((....)).))))))..((((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))). ( -32.60) >consensus UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUGUCGGUGUCGCUCAGAUUCACAGGAGUGAAACAAAUCCCUUUACUCAUAAUUCAUUAAUUUCUGUAUUUAGAUUUGAGCGC ....(((((((((((((((......)))))).....)))))))))..((((((((((.(((((((((((.........))))))).(((....)))....)))).....)))))))))). (-33.82 = -34.42 + 0.60)

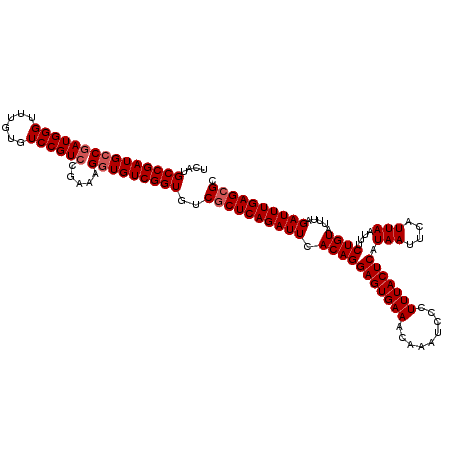
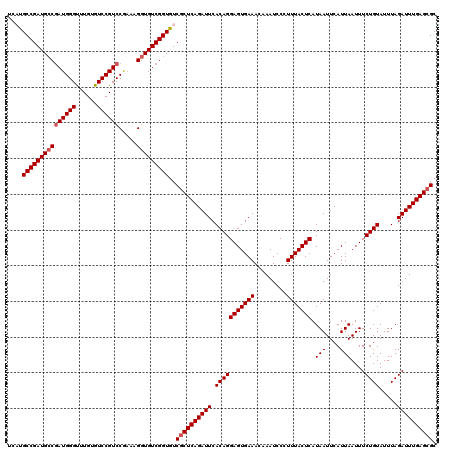
| Location | 1,670,527 – 1,670,624 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
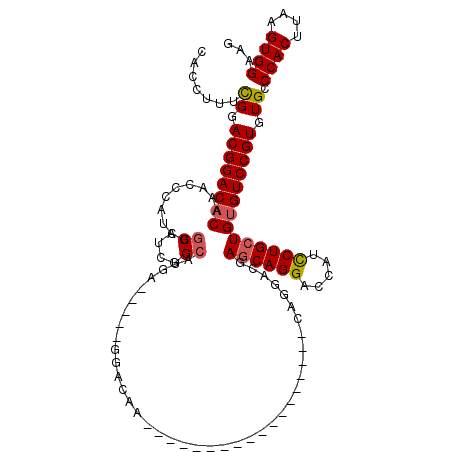
>X_DroMel_CAF1 1670527 97 + 22224390 CACCUUUCGGACGGACACAAACCCAUCGGCAUCGGCAUGA-----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAG .......((.((((((((....((.(((((....)).)))-----))....------------------.......((((((.....)))))))))))))).)).((((....))))... ( -35.70) >DroSec_CAF1 4594 97 + 1 CACCUUUUGGACGGACACAAACCCAUCGGCAUCGGCAUGA-----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAG ..((....))((((((((....((.(((((....)).)))-----))....------------------.......((((((.....))))))))))))))....((((....))))... ( -35.30) >DroSim_CAF1 5446 97 + 1 CACCUUUCGGACGGACACAAACCCAUCGGCAUCGGCAUGA-----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAG .......((.((((((((....((.(((((....)).)))-----))....------------------.......((((((.....)))))))))))))).)).((((....))))... ( -35.70) >DroEre_CAF1 8226 113 + 1 CACCUUUCGGACGGACCCAAGCCCAUCGGCAUCGGCA--A-----CGACAACAGGACGAGCAGGACGGGCAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAG ..(((((((..(........(((....))).(((...--.-----))).....)..)))).)))..((((((((((((((((.....)))))).)))))...)))))............. ( -44.80) >DroYak_CAF1 5624 114 + 1 CACCUUACGGACGGACACAAACCCAU-GCCAUCGGCA--ACCCAACGACAACG---AGAACAGGACGAGCAGGACGAACAGGACCAUUCUGAUGUGUCCGUGUGCCCACUUAAGUGGAAG .......((.(((((((((...((.(-(((.(((...--.)....((....))---.......)).).))))).....(((((...))))).))))))))).)).((((....))))... ( -31.40) >consensus CACCUUUCGGACGGACACAAACCCAUCGGCAUCGGCAUGA_____GGACAA__________________CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAG .......((.((((((((..........((....))........................................((((((.....)))))))))))))).)).((((....))))... (-27.90 = -28.18 + 0.28)

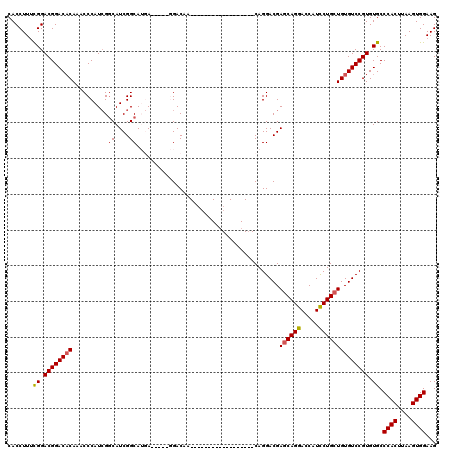
| Location | 1,670,527 – 1,670,624 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -33.20 |
| Energy contribution | -33.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
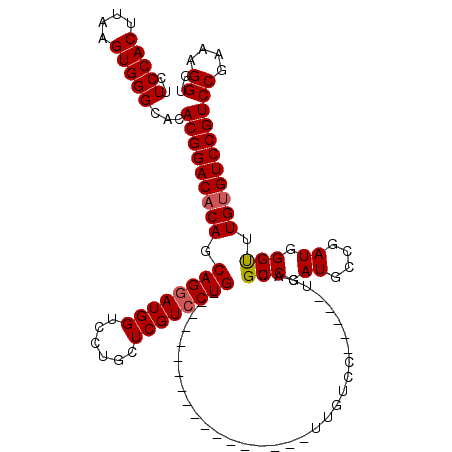
>X_DroMel_CAF1 1670527 97 - 22224390 CUUCCACUUAAGUGGGCACACGGACACAGCAGGAUGGUCCUGCUCGUCCUG------------------UUGUCC-----UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUG ..(((((....)))))...((((((((((((((.....)))))....((((------------------(((.(.-----((.....)).).)))))))..)))))))))((....)).. ( -39.20) >DroSec_CAF1 4594 97 - 1 CUUCCACUUAAGUGGGCACACGGACACAGCAGGAUGGUCCUGCUCGUCCUG------------------UUGUCC-----UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCAAAAGGUG ....(((((...(((((((((((((((((((((((((......))))))))------------------))))((-----((..((....)).)).))))))))))..)))))..))))) ( -38.00) >DroSim_CAF1 5446 97 - 1 CUUCCACUUAAGUGGGCACACGGACACAGCAGGAUGGUCCUGCUCGUCCUG------------------UUGUCC-----UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUG ..(((((....)))))...((((((((((((((.....)))))....((((------------------(((.(.-----((.....)).).)))))))..)))))))))((....)).. ( -39.20) >DroEre_CAF1 8226 113 - 1 CUUCCACUUAAGUGGGCACACGGACACAGCAGGAUGGUCCUGCUCGUCCUGCCCGUCCUGCUCGUCCUGUUGUCG-----U--UGCCGAUGCCGAUGGGCUUGGGUCCGUCCGAAAGGUG ..(((((....)))))...((((((...(((((((((......)))))))))(((....(((((((..(..((((-----.--...))))..)))))))).)))))))))((....)).. ( -46.30) >DroYak_CAF1 5624 114 - 1 CUUCCACUUAAGUGGGCACACGGACACAUCAGAAUGGUCCUGUUCGUCCUGCUCGUCCUGUUCU---CGUUGUCGUUGGGU--UGCCGAUGGC-AUGGGUUUGUGUCCGUCCGUAAGGUG ..(((((....)))))...(((((((((.(((.((((......)))).)))...........((---(((.((((((((..--..))))))))-)))))..)))))))))((....)).. ( -40.10) >consensus CUUCCACUUAAGUGGGCACACGGACACAGCAGGAUGGUCCUGCUCGUCCUG__________________UUGUCC_____UCAUGCCGAUGCCGAUGGGUUUGUGUCCGUCCGAAAGGUG ..(((((....)))))...(((((((((.((((((((......)))))))).................................(((.((....)).))).)))))))))((....)).. (-33.20 = -33.44 + 0.24)

| Location | 1,670,567 – 1,670,664 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1670567 97 + 22224390 -----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAACCAUACCUGAACGUGUGGAGGUUU -----(.(((.------------------..(((((((((((.....)))))).))))).))).)((((....))))...................((((((......))))))...... ( -33.40) >DroSec_CAF1 4634 97 + 1 -----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAACCAUACCUGAGCGUGUGGAGGUUU -----(.(((.------------------..(((((((((((.....)))))).))))).))).)((((....))))...................((((((......))))))...... ( -33.40) >DroSim_CAF1 5486 97 + 1 -----GGACAA------------------CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAACCAUACCUGAGCGUGUGGAGGUUU -----(.(((.------------------..(((((((((((.....)))))).))))).))).)((((....))))...................((((((......))))))...... ( -33.40) >DroEre_CAF1 8264 115 + 1 -----CGACAACAGGACGAGCAGGACGGGCAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAAGCAUACCUGAGCGUGUGGAGGUUU -----.(((....(.(((..((((..((((((((((((((((.....)))))).)))))...)))))(((...(((........)))....))).......))))..))).)....))). ( -41.80) >DroYak_CAF1 5661 117 + 1 CCCAACGACAACG---AGAACAGGACGAGCAGGACGAACAGGACCAUUCUGAUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAAGCAUACCUGAGCGUGUGGAGAUUU .((((((.(....---......(((((..(((((.(........).)))))...)))))((((((((((....))))...((..........))..))))))....)))).)))...... ( -28.80) >consensus _____GGACAA__________________CAGGACGAGCAGGACCAUCCUGCUGUGUCCGUGUGCCCACUUAAGUGGAAGACGACAUAAAUAGUAACCAUACCUGAGCGUGUGGAGGUUU .......((......................(((((((((((.....)))))).)))))((.(..((((....)))).).))..........))..((((((......))))))...... (-27.88 = -28.32 + 0.44)

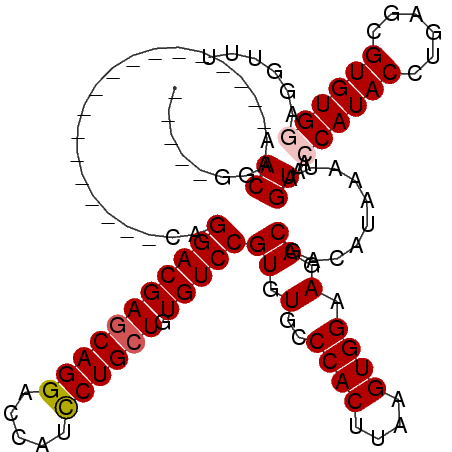
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:06 2006