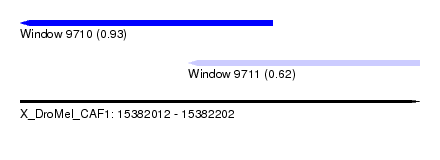
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,382,012 – 15,382,202 |
| Length | 190 |
| Max. P | 0.931771 |

| Location | 15,382,012 – 15,382,132 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -28.83 |
| Energy contribution | -31.82 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382012 120 - 22224390 GAUGCUGAAUCUGAUGCUGAUGCUGAAGCUGUAGCUGAAACUGAAGCUAAGGCUGACUCAAGGUGACGUAAUGGCAACGCCUAAAGUGGCUUUUAUACGCUCCACCAGAUCAAGUAUCAA ((((((..(((((..((....))(((((((.(((((........))))).))))...))).((.(.((((..(....)(((......))).....))))).))..)))))..)))))).. ( -38.70) >DroSec_CAF1 24210 120 - 1 GAUGCUGAAUCUGAUGCUGAUGCCGAAGCUGUAGCUGAAACUGAAGCUAAGGCUGACUCAAGGUGACGUAAUGGCAACGCCUAAAGUGGCUUUUAUACGCUCCACCAGAUCAAGUAUCAA ((((((..(((((.((..(.(((((.((((.(((((........))))).)))).......(....)....)))))..(((......))).........)..)).)))))..)))))).. ( -39.00) >DroEre_CAF1 21808 114 - 1 GAUGCUGAAUCUGAUGCUGA------AGCUGAAGCUGAAACUGAAGCUAAGGCUGACUCAAGGUGACGUAAUGGCAACGCCUAAAGUGGCUUUUAUACGCUCCACCAGAUCAAGUAUCAA ((((((..(((((.((....------((((..((((........))))..))))....)).((.(.((((..(....)(((......))).....))))).))..)))))..)))))).. ( -36.90) >DroYak_CAF1 14036 102 - 1 ------GAAUCUGAUGCUGA------A------GCUGAAACUGAAGCUAAGGCUGACUCAAGGUGACGUAAUGGCAACGCCUAAAGUGGCUUUUAUACGCUCCACCAGAUCAAGUAUCAA ------.....(((((((..------.------.(((....((.(((.((((((.(((..(((((..((....))..)))))..))))))))).....))).)).)))....))))))). ( -28.00) >consensus GAUGCUGAAUCUGAUGCUGA______AGCUGUAGCUGAAACUGAAGCUAAGGCUGACUCAAGGUGACGUAAUGGCAACGCCUAAAGUGGCUUUUAUACGCUCCACCAGAUCAAGUAUCAA ((((((..(((((....(((......((((.(((((........))))).))))...))).((.(.((((..(....)(((......))).....))))).))..)))))..)))))).. (-28.83 = -31.82 + 3.00)

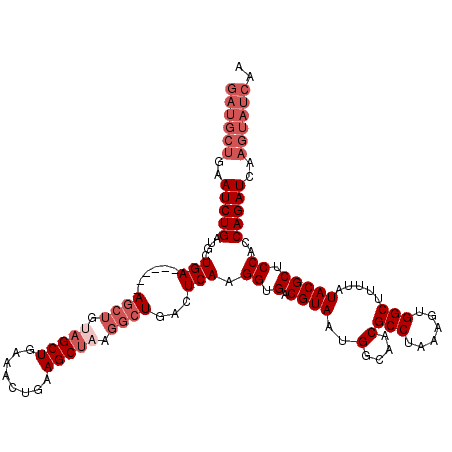
| Location | 15,382,092 – 15,382,202 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -16.21 |
| Energy contribution | -17.77 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15382092 110 - 22224390 ----------CUGUAUCUGUGGUUAUGACGCCAAAGCUAUAGCUAAAGCUACAGCUAAAACUGAAUCUGAAUCUGAAUCUGAUGCUGAAUCUGAUGCUGAUGCUGAAGCUGUAGCUGAAA ----------.........((((......)))).((((((((((..(((..((((.......((.((...(((.......)))...)).))....))))..)))..)))))))))).... ( -31.60) >DroSec_CAF1 24290 108 - 1 GUAGCUACAGCUGUUACUG------UGACGCCAAAGCUAAAGCUAACGCUACAGCUAAAACUGAAUCU------GAAUCUGAUGCUGAAUCUGAUGCUGAUGCCGAAGCUGUAGCUGAAA .((((((((((((((....------.)))((...((((..(((....)))..))))........(((.------..(((.(((.....))).)))...)))))...)))))))))))... ( -33.10) >DroEre_CAF1 21888 102 - 1 GUAGCUACAGCUGUUACUCUGGCUAUGACGCCAAAG------CUAACGCUACAGCUAAAACUGAAUCU------GAAUCUGAUGCUGAAUCUGAUGCUGA------AGCUGAAGCUGAAA .(((((.(((((((((((.((((......)))).))------.))))....((((.....(.......------).(((.(((.....))).))))))).------))))).)))))... ( -29.70) >consensus GUAGCUACAGCUGUUACUGUGG_UAUGACGCCAAAGCUA_AGCUAACGCUACAGCUAAAACUGAAUCU______GAAUCUGAUGCUGAAUCUGAUGCUGAUGC_GAAGCUGUAGCUGAAA ................((.((((......)))).))...........(((((((((.........((.......))(((.(((.....))).)))...........)))))))))..... (-16.21 = -17.77 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:08 2006