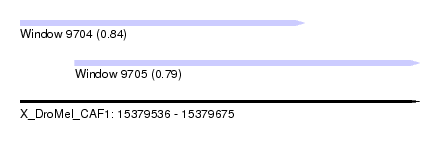
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,379,536 – 15,379,675 |
| Length | 139 |
| Max. P | 0.835307 |

| Location | 15,379,536 – 15,379,635 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15379536 99 + 22224390 ---------CAGGGGUGCAGGGUGAUCUCCGGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAU ---------.(((((((..(((.....)))((.((((.(((((...(((....)))))))).)))))).((((....))))..)))))))........(((....))) ( -33.00) >DroSim_CAF1 14891 99 + 1 ---------CAGGGGUGCAGGGUGAUCCCCGGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAU ---------.(((((((..(((.....)))((.((((.(((((...(((....)))))))).)))))).((((....))))..)))))))........(((....))) ( -35.50) >DroEre_CAF1 19514 93 + 1 ---------------UACAGGGUGAUCGCCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGUGCGGUAAUUACCCCUGAAUGGGAAUGGAAACAU ---------------..(((((((((..((.(..(((.(((((...(((....)))))))).)))..)..((...))))..)))).))))).......(((....))) ( -31.20) >DroYak_CAF1 11620 108 + 1 AUACAUGCACGGGGGUGCAGGGUGAUCUCCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGUGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAU .....((((.(((((((((.(((((((......)))).(((((...(((....)))))))).........))).))).......))))))...)))).(((....))) ( -34.41) >consensus _________CAGGGGUGCAGGGUGAUCUCCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAU ...............(((((((((((..((.(..(((.(((((...(((....)))))))).)))..)..((...))))..))))))).....)))).(((....))) (-28.92 = -29.18 + 0.25)

| Location | 15,379,555 – 15,379,675 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -37.69 |
| Energy contribution | -37.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15379555 120 + 22224390 CCGGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAUUCUCAUUUUUAUGAGGCACACGCUGGCCGGCGAGACAAUG ..((.((((.(((((...(((....)))))))).)))))).(((((.(((((....)))...(((((.(((((....))))).)))))......(((....))).)))))))........ ( -39.30) >DroSim_CAF1 14910 120 + 1 CCGGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAUUCUCAUUUUUAUGAGGCACACGCUGGCCGGCGAGACAAUG ..((.((((.(((((...(((....)))))))).)))))).(((((.(((((....)))...(((((.(((((....))))).)))))......(((....))).)))))))........ ( -39.30) >DroEre_CAF1 19527 120 + 1 CCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGUGCGGUAAUUACCCCUGAAUGGGAAUGGAAACAUUCUCAUUUUUAUGAGGCACACGUUGGCCGGCGAGACAAUG ...(..(((.(((((...(((....)))))))).)))..).(((((.(((((....)))((((((((((((((....))))))))))).....))).........)))))))........ ( -41.60) >DroYak_CAF1 11648 120 + 1 CCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGUGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAUUCUCAUUUUUAUGAGGCACACGCUGGCCGGCGAGACAAUG (((((((((.(((((...(((....)))))))).)))........(((((((....)))...(((((.(((((....))))).))))).......)))).)))))).............. ( -38.00) >consensus CCAGUGAUCCCUGGGUGAAUCAAAUGAUCCCAGUGAUUCCAUGCCGCGCGGUAAUUACCCCUAAAUGCGAAUGGAAACAUUCUCAUUUUUAUGAGGCACACGCUGGCCGGCGAGACAAUG ...(..(((.(((((...(((....)))))))).)))..).(((((.(((((....)))((((((((.(((((....))))).))))).....))).........)))))))........ (-37.69 = -37.50 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:03 2006