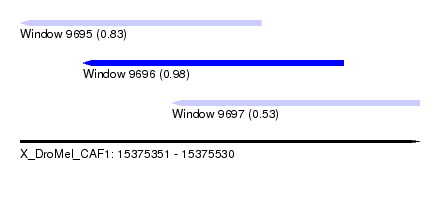
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,375,351 – 15,375,530 |
| Length | 179 |
| Max. P | 0.975592 |

| Location | 15,375,351 – 15,375,459 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15375351 108 - 22224390 UCAUGAGCACAUCAUCAUUAUCAUUGUGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGCUUCCCGUU---GUCCACGGGGCUUCU--------- ......((((((.(((((......((....)).....))).)).))))))((((((.......((((((.....)))).))((((........---)))).))))))....--------- ( -30.90) >DroSec_CAF1 17380 111 - 1 UCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGCUUCCCGUU---GUCCACAGGCUUGCUAAG------ .....((((.............(((((((((((...((.....)).))).))))))))..........((((((.((((((.((....)).))---)))))))))).))))...------ ( -35.60) >DroSim_CAF1 10741 111 - 1 UCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGCUUCCCGUU---GUCCACGGGGUUGCUAGG------ .....((((.............(((((((((((...((.....)).))).))))))))......((((((.....((((((.((....)).))---)))))))))).))))...------ ( -35.60) >DroEre_CAF1 15581 117 - 1 UCAUGAGCACAUCGUCAUUAUCAUUGUGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGUUUCCCGUU---GUCCACGGGGUUUCGCGAGGUUCC ....((((...(((((((.......)))))................(((.((((((.....((........))..((((((.((....)).))---)))).))))))..))))).)))). ( -34.60) >DroYak_CAF1 7414 120 - 1 UCAUGAGCACAUCAUCAUUAUCAUCGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGCUUCCCGUUCCCGUCCACGGGGUUGCGGAAGGUUCC ....((((..............(((((((((((...((.....)).))).))))))))..((.((((((.....)))).)))).(((((((..((((....))))..)).))))))))). ( -43.60) >consensus UCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACGAGGGCUUCCCGUU___GUCCACGGGGUUGCUAAA______ ....((((..............(((((((((((...((.....)).))).))))))))..((.((((((.....)))).))))))))(((((........)))))............... (-28.70 = -28.98 + 0.28)



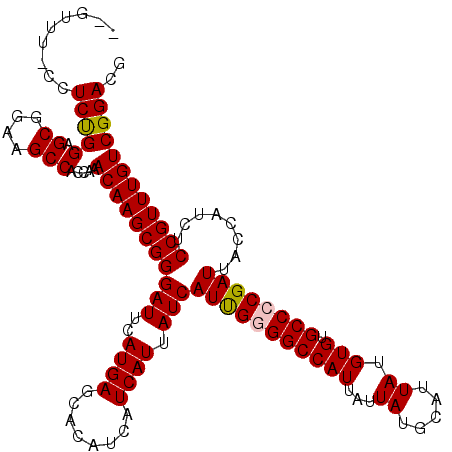
| Location | 15,375,379 – 15,375,496 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -32.86 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15375379 117 - 22224390 --GGUU-UCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGUGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG --((((-(((.......))))))).((..(((((((((((..((((........)))).)))((((.((((((...((.....)).))).))).))))........)))))))).))... ( -30.40) >DroSec_CAF1 17411 118 - 1 --UUUUCCCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG --...(((...(((.((....))..))).(((((((((((..((((........)))).)))(((((((((((...((.....)).))).))))))))........)))))))).))).. ( -37.00) >DroSim_CAF1 10772 117 - 1 --UUUU-CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG --...(-((..(((.((....))..))).(((((((((((..((((........)))).)))(((((((((((...((.....)).))).))))))))........)))))))).))).. ( -37.40) >DroEre_CAF1 15618 115 - 1 ----UU-CCUCCGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCGUCAUUAUCAUUGUGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG ----..-..(((((.((....))..))..(((((((((((((..(.((((((.(((((.......)))))(((....)))......)))))))..)).....))).)))))))).))).. ( -30.50) >DroYak_CAF1 7454 119 - 1 UUGUUU-CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUCGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG .....(-((..(((.((....))..))).(((((((((((..((((........)))).)))(((((((((((...((.....)).))).))))))))........)))))))).))).. ( -39.50) >consensus __GUUU_CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUGCAUUAUGUGUGCCCCGAUUACCAUCUCCGUUUGUCGGACG .........(((((.((....))).....(((((((((((..((((........)))).)))(((((((((((...((.....)).))).))))))))........)))))))))))).. (-32.86 = -32.94 + 0.08)


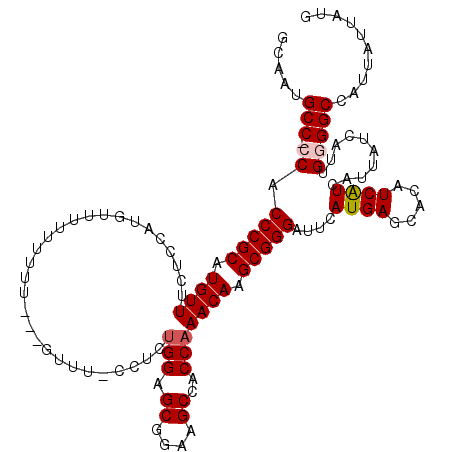
| Location | 15,375,419 – 15,375,530 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.61 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15375419 111 - 22224390 GCAAUGCC-CCACCCGCAUGUUUUUU----UUUUCCUCU---GGUU-UCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGUGGCCAUUAUUAUG ((((((..-...(((((.(((((...----((((((((.---.(..-...)..))).))))).....))))).)))))....((((.....))))......))))))............. ( -28.50) >DroSec_CAF1 17451 117 - 1 GCAAUGCCCCCACCCGCAUGUUUCUCUAUGUUUUUUUUU---UUUUCCCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUG ..((((.((((((((((.(((((................---...(((....)))((....))....))))).)))))....((((.....)))).........))))).))))...... ( -32.50) >DroSim_CAF1 10812 116 - 1 GCAAUGCCCCCACCCGCAUGUUUCUCUAUGUUUUUUUUU---UUUU-CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUG ..((((.((((((((((.(((((................---...(-((...)))((....))....))))).)))))....((((.....)))).........))))).))))...... ( -31.60) >DroEre_CAF1 15658 110 - 1 GCGCUGCC-CCGCCCGCAUGUUGCCCCAUGUUUUUU--------UU-CCUCCGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCGUCAUUAUCAUUGUGGCCAUUAUUAUG (.(((...-...(((((.((((.......(((((..--------((-((...))))..))))).....)))).))))).......))).)...(((((.......))))).......... ( -26.62) >DroYak_CAF1 7494 118 - 1 GCAAUGCC-CCACCCGCAUGUUUCUCCAUGCUUUUUUUUUUUGUUU-CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUCGGGGCCAUUAUUAUG .....(((-((.((((((((......)))))........((((((.-....(((.((....))..))))))))).)))....((((.....))))..........))))).......... ( -32.10) >consensus GCAAUGCC_CCACCCGCAUGUUUCUCCAUGUUUUUUUUU___GUUU_CCUCUGGAGCGGAAGCCACCAAACAAGCGGGAUUCAUGAGCACAUCAUCAUUAUCAUUGGGGCCAUUAUUAUG .....(((.((.(((((.((((.............................(((.((....))..))))))).)))))....((((.....))))..........))))).......... (-22.17 = -22.61 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:55 2006