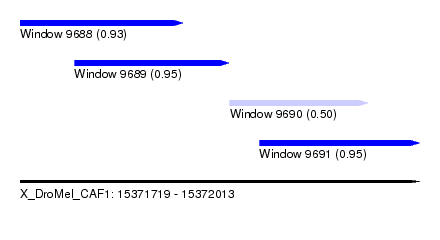
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,371,719 – 15,372,013 |
| Length | 294 |
| Max. P | 0.954959 |

| Location | 15,371,719 – 15,371,839 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -40.64 |
| Consensus MFE | -29.38 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15371719 120 + 22224390 AUUUUUAUCGACGAAAAUGUAGUAGCUAUUAAACAACAUUUUCAUUGCGCUUUUCUCGCAGUGCAGGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACUCGGACGUG .......((((.((((((((.((.........)).))))))))..((((.......))))(..(((((((...((((((((......))))))))...)))))))..)...))))..... ( -43.80) >DroSec_CAF1 13709 120 + 1 AUUUUUAUCGGCGAAAAUGUAGUGGCUAUUAAACUACAUUUUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACGCGGACUUG .........((((((((((((((.........)))))))))))...)))....((.(((..(((((((((...((((((((......))))))))...))))..)))))..))))).... ( -45.80) >DroSim_CAF1 6874 120 + 1 AUUUUUAUCGGCGAAAAUAUAGUGGCUAUUAAACUACAUUUUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACGCGGACGUG .........(((((((((.((((.........)))).))))))...)))....((.(((..(((((((((...((((((((......))))))))...))))..)))))..))))).... ( -39.80) >DroEre_CAF1 11811 120 + 1 AUUUUUAUCGGUGGAAAUAUGGUCGCUAUUAAACUACAUUUUCAUAGCUCUUUUCUCACAGUGCAGGAGUUUUGGAGCGCUGGAAGUUGCGUUCCUUGGCUUCUGUGCAACUCGGACGUG .......((((((((((...((..(((((.(((......))).))))).)))))).))).(..((((((((..(((((((........)))))))..))))))))..)....)))..... ( -39.60) >DroYak_CAF1 3783 120 + 1 AUUUUUAUUGGUGAGAAUAGAGUCGCUAUUAAACUACAUUUUCAUGGCUAUUUUCUCGUAGUGCAUUAGCUUUGGAGCGCUGGAAGUUGCGUUCUUUGGCUUCUGUGCAACUUGGACGUG ...((((.((((((........)))))).))))...(((.((((.((((((......)))))((((.((((..(((((((........)))))))..))))...))))..).)))).))) ( -34.20) >consensus AUUUUUAUCGGCGAAAAUAUAGUCGCUAUUAAACUACAUUUUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACUCGGACGUG .......(((..((((((.((((.........)))).)))))).................(..((.((((...(((((((........)))))))...)))).))..)....)))..... (-29.38 = -28.90 + -0.48)


| Location | 15,371,759 – 15,371,873 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -43.54 |
| Consensus MFE | -37.14 |
| Energy contribution | -36.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15371759 114 + 22224390 UUCAUUGCGCUUUUCUCGCAGUGCAGGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACUCGGACGUGGCUGAACAUGUGGAAUGUAUGGAUGUGCCUGUG------C ..(((.((((..(((..((((..(((((((...((((((((......))))))))...)))))))..).......(((((......)))))....)))..))).))))..)))------. ( -45.20) >DroSec_CAF1 13749 119 + 1 UUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACGCGGACUUGGACGAACAUGUGG-AUGUAUAGAUGUGCCUGUGGACGUGG .......((((..((.(((((.((((((((...((((((((......))))))))...)))((((((((.((((...((....))...)))).-.)))))))))))))))))))).)))) ( -43.80) >DroSim_CAF1 6914 119 + 1 UUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACGCGGACGUGGCGGAACAUGUGG-AUGUAUGGAUGUGCCUGUGGACGUGG ....(((((((..((.(((..(((((((((...((((((((......))))))))...))))..)))))..))))).)))))))..(((((..-((((((....))))..))..))))). ( -43.90) >DroEre_CAF1 11851 119 + 1 UUCAUAGCUCUUUUCUCACAGUGCAGGAGUUUUGGAGCGCUGGAAGUUGCGUUCCUUGGCUUCUGUGCAACUCGGACGUGAAUGAGCACGUGG-AUGUAUGGAUGUGCCUGUGAAUAUGG ((((((((.(..(((..((((..((((((((..(((((((........)))))))..))))))))..).......(((((......)))))..-.)))..))).).).)))))))..... ( -44.80) >DroYak_CAF1 3823 119 + 1 UUCAUGGCUAUUUUCUCGUAGUGCAUUAGCUUUGGAGCGCUGGAAGUUGCGUUCUUUGGCUUCUGUGCAACUUGGACGUGAAUGAGCACGUGG-AUGUAUGGAUGUGCCUGUGGACGUGG (((((((((...(((.(((..(((((.((((..(((((((........)))))))..))))...)))))......))).)))..))).)))))-).......((((.((...)))))).. ( -40.00) >consensus UUCAUUGCUAUUUUCUCGCAGUGCACGAGCAUUGGAGCGCUGGAAGUGGCGUUCCUUGGCUUCUGUGCAACUCGGACGUGGAUGAACAUGUGG_AUGUAUGGAUGUGCCUGUGGACGUGG ...............((((((.((((((((...(((((((........)))))))...)))((((((((......(((((......)))))....)))))))))))))))))))...... (-37.14 = -36.98 + -0.16)

| Location | 15,371,873 – 15,371,975 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15371873 102 + 22224390 AUG------UG------------GCCAUGGUCGACCCAUUUAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGU ...------((------------((((((((...........((((((((((((((((.....))))))))).....)))))))((.....))....)))))).))))............ ( -36.50) >DroSec_CAF1 13868 102 + 1 AUG------UG------------GACAUGGUCGACCCAUUUAGGAGUGCUCGCUAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAACACAAAGU .((------((------------...((((.....))))...(((((((..((((((..(....)..))))))....))))))).((((.(((((.....))))).))))..)))).... ( -30.50) >DroSim_CAF1 7033 102 + 1 AUG------UG------------GACAUGGUCGACCCAUUUAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGU ..(------..------------(((((((.....))))...((((((((((((((((.....))))))))).....))))))))))..).((((.....))))((.....))....... ( -36.10) >DroEre_CAF1 11970 108 + 1 AUGUGAGUGUG------------GACAUGGUCGACCCACUUAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUCGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGGCAAAAGCACAAAGU .((((((((.(------------(.(......).))))))..((((((((((((((((.....))))))))).....))))))).((((((((((.....))))))))))..)))).... ( -44.90) >DroYak_CAF1 3942 120 + 1 AUGUGGAUGUGGAUGUGGAUACGGACAUGGUCGACCCACUUAGGAGUGCUCGCGAUGGAGACGCCGGCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGGCAAAAGCACAAAGU ..((((..((.((((((........))).))).))))))......(((((...(((((((.(((.((((....))))))))))))))((((((((.....))))))))..)))))..... ( -43.70) >consensus AUG______UG____________GACAUGGUCGACCCAUUUAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGU ...........................(((.....)))....((((((((((((((((.....))))))))).....))))))).((((.(((((.....))))).)))).......... (-29.54 = -29.94 + 0.40)


| Location | 15,371,895 – 15,372,013 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -46.18 |
| Consensus MFE | -37.64 |
| Energy contribution | -37.56 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15371895 118 + 22224390 UAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGUCCUGCCAGGACGCGAAAUUGCUAGAUGGAGUGGG--AAAG .......((.((((((((.....))))))))))..((.((((((((((((.((((.....))))))....((((....((((.....)))).......)))))))))))))).)--)... ( -41.60) >DroSec_CAF1 13890 117 + 1 UAGGAGUGCUCGCUAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAACACAAAGUCCUGCCAGGACGCGAAAUGGUUCGAUGGAGUGGG--A-AG ..(((((((..((((((..(....)..))))))....))))))).((..((.(((.((((((.(((............((((.....)))))))..))))))..))).))..))--.-.. ( -36.20) >DroSim_CAF1 7055 117 + 1 UAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGUCCUGCCAGGACGCGAAAUGGUUAGAUGGAGUGGG--A-AG ..((((((((((((((((.....))))))))).....))))))).((..((.(((.((((((((((.....)).....((((.....))))))...))))))..))).))..))--.-.. ( -43.40) >DroEre_CAF1 11998 117 + 1 UAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUCGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGGCAAAAGCACAAAGUCCUGCCAGGACGCGAAAUGGCAGGACGGGGCGGG--U-AG ......((((((((((((..(((....)))..)))))))(((((.((((((((((.....))))))))))........(((((((((..........)))))))))))))))))--)-). ( -57.80) >DroYak_CAF1 3982 119 + 1 UAGGAGUGCUCGCGAUGGAGACGCCGGCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGGCAAAAGCACAAAGUCCUGCCAGGACGCAAAAUGGCAGGAUGGAGUGGGGUU-AG .......((((((((..(..(((....)))..)..))))(((((.((((((((((.....))))))))))........(((((((((..........)))))))))))))).)))).-.. ( -51.90) >consensus UAGGAGUGCUCGCGAUGGAGACGCCGUCGUGGUUGUCGCGCUCCGUUUGCUGCAUAAAUCAUGCGCCAAAAGCACAAAGUCCUGCCAGGACGCGAAAUGGCUAGAUGGAGUGGG__A_AG ...((..((.((((((((.....))))))))))..)).(((((((((((((((((.....))))..............((((.....)))).......))).))))))))))........ (-37.64 = -37.56 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:50 2006