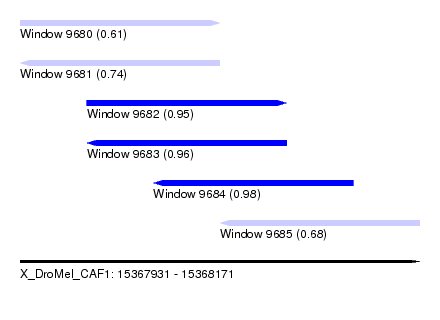
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,367,931 – 15,368,171 |
| Length | 240 |
| Max. P | 0.984428 |

| Location | 15,367,931 – 15,368,051 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -24.89 |
| Energy contribution | -24.82 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367931 120 + 22224390 CAUUUUUGCCUAUCACAGCAUAUAUGAAAUUAAUAACCUUUGCCAACUUUCUGGCCAAAGUGUGCUUUUCUCGGCAAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUU ....((((((......(((((((((.......)))..((((((((......))).)))))))))))......))))))(((((....((((...)))).)))))...(((....)))... ( -29.90) >DroSec_CAF1 745 120 + 1 CAUUUUUGGCUAUCACCGCAUAUACGAAAUCAAUAACCUUUGCCAACUUUUUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUU ................(((((((.(((........(((((((((((....)))).))))).)).......)))((.(.(((((....((((...)))).))))).).))))))))).... ( -32.46) >DroSim_CAF1 3250 120 + 1 CAUUUUUGGCUAUCACCGCAUAUACGAAAUUAAUAACCUUUGCCAACUUUUUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUU ................(((((((.(((........(((((((((((....)))).))))).)).......)))((.(.(((((....((((...)))).))))).).))))))))).... ( -32.46) >DroEre_CAF1 7885 120 + 1 CCUGGUUGAGUAUGACAGCAUACAUGAAAUUAAAAACCACUUCAAACUUUCCGGCCAAAGUGUGCGUUUCUCGGCCAAUUGUGCCAUCAGCUCUGCUGUCACACCUGGCGUAUGCGUAUU ..((((((.(((((....))))).((((............)))).......))))))....((((((....((.(((..((((....((((...)))).))))..)))))...)))))). ( -29.50) >consensus CAUUUUUGGCUAUCACAGCAUAUACGAAAUUAAUAACCUUUGCCAACUUUCUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUU .................((((((.......................(.....)((((..(((((........((((.....))))..((((...)))).))))).))))))))))..... (-24.89 = -24.82 + -0.06)


| Location | 15,367,931 – 15,368,051 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -26.09 |
| Energy contribution | -26.47 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367931 120 - 22224390 AAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUUGCCGAGAAAAGCACACUUUGGCCAGAAAGUUGGCAAAGGUUAUUAAUUUCAUAUAUGCUGUGAUAGGCAAAAAUG ...........(((...(((.((((((..(((..(((((.....))))).....)))..(((((.(((((....))))).)))))...............)))))).))))))....... ( -32.60) >DroSec_CAF1 745 120 - 1 AAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCAAAGGUUAUUGAUUUCGUAUAUGCGGUGAUAGCCAAAAAUG .....(((((.((.(((((((.((((...))))....))))))).))(((((.......(((((.(((((....))))).)))))......)))))..)))))(((....)))....... ( -35.02) >DroSim_CAF1 3250 120 - 1 AAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCAAAGGUUAUUAAUUUCGUAUAUGCGGUGAUAGCCAAAAAUG .....(((((.((.(((((((.((((...))))....))))))).))(((((.......(((((.(((((....))))).)))))......)))))..)))))(((....)))....... ( -35.02) >DroEre_CAF1 7885 120 - 1 AAUACGCAUACGCCAGGUGUGACAGCAGAGCUGAUGGCACAAUUGGCCGAGAAACGCACACUUUGGCCGGAAAGUUUGAAGUGGUUUUUAAUUUCAUGUAUGCUGUCAUACUCAACCAGG .....((....))..(((((((((((((((((..((((.(((.((((........)).))..)))))))...)))))(((((........))))).....))))))))))))........ ( -35.80) >consensus AAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCAAAGGUUAUUAAUUUCAUAUAUGCGGUGAUAGCCAAAAAUG .....(((((.((.(((((((.((((...))))....))))))).)).((((.......(((((.(((((....))))).)))))......))))...)))))................. (-26.09 = -26.47 + 0.38)

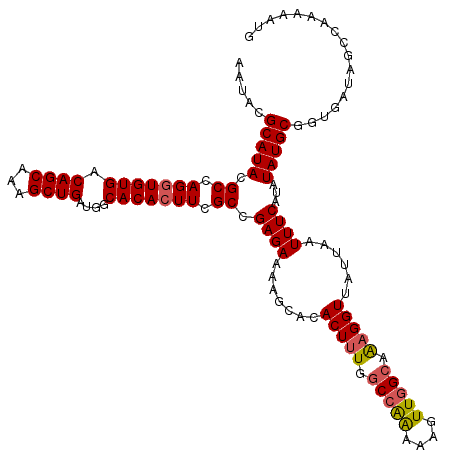

| Location | 15,367,971 – 15,368,091 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -33.44 |
| Energy contribution | -34.75 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367971 120 + 22224390 UGCCAACUUUCUGGCCAAAGUGUGCUUUUCUCGGCAAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUUUAUUUGCUAUAUGUGCGCAUUUUAAUAGGCCACUUUGCGU .((((......))))(((((((.((((......((...(((((....((((...)))).)))))...))..((((((((.(((.....))).))))))))......)))))))))))... ( -38.20) >DroSec_CAF1 785 120 + 1 UGCCAACUUUUUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUUUAUUUGCCAUAUGUGCGCAUUUUAACAGGGCACUUUGCGU .(((((....)))))((((((((........(((((((((.........))))))))).....((((....((((((((.(((.....))).)))))))).....))))))))))))... ( -41.40) >DroSim_CAF1 3290 120 + 1 UGCCAACUUUUUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUUUAUUUGCCAUAUGUGCGCAUUUUAACAGGGCACUUUGCGU .(((((....)))))((((((((........(((((((((.........))))))))).....((((....((((((((.(((.....))).)))))))).....))))))))))))... ( -41.40) >DroEre_CAF1 7925 120 + 1 UUCAAACUUUCCGGCCAAAGUGUGCGUUUCUCGGCCAAUUGUGCCAUCAGCUCUGCUGUCACACCUGGCGUAUGCGUAUUUAUUUGCCAUAUGUGCGCAUUUUGACAGGACACUUUGCGU ..((((.(.(((.(.(((((((((((.......((((..((((....((((...)))).))))..))))(((((.(((......)))))))).))))))))))).).))).).))))... ( -37.30) >consensus UGCCAACUUUCUGGCCAAAGUGUGCUUUUCUCGGCGAAGUGUGCCAUCAGCUUUGCUGUCACACCUGGCGUAUGCGUAUUUAUUUGCCAUAUGUGCGCAUUUUAACAGGGCACUUUGCGU .((((......))))((((((((........(((((((((.........))))))))).....((((....((((((((.(((.....))).)))))))).....))))))))))))... (-33.44 = -34.75 + 1.31)

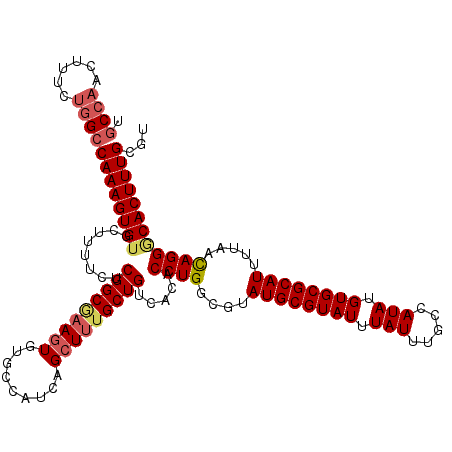
| Location | 15,367,971 – 15,368,091 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367971 120 - 22224390 ACGCAAAGUGGCCUAUUAAAAUGCGCACAUAUAGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUUGCCGAGAAAAGCACACUUUGGCCAGAAAGUUGGCA ...(((((((((........(((((....(((.....)))....)))))..((.(((((((.((((...))))....))))))).))........)).)))))))(((((....))))). ( -35.10) >DroSec_CAF1 785 120 - 1 ACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCA ...(((((((...........(((.(((((.((((................)))).)))))...)))..(((..((((.......)))).....))).)))))))(((((....))))). ( -36.29) >DroSim_CAF1 3290 120 - 1 ACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCA ...(((((((...........(((.(((((.((((................)))).)))))...)))..(((..((((.......)))).....))).)))))))(((((....))))). ( -36.29) >DroEre_CAF1 7925 120 - 1 ACGCAAAGUGUCCUGUCAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAGAGCUGAUGGCACAAUUGGCCGAGAAACGCACACUUUGGCCGGAAAGUUUGAA ...((((.(.(((.((((((.(((((((((.((((................)))).))))).((((...)))).((((.(....))))).....))))...)))))).))).).)))).. ( -35.39) >consensus ACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCACACUUCGCCGAGAAAAGCACACUUUGGCCAAAAAGUUGGCA ...(((((((...........(((.(((((.((((................)))).)))))...)))..(((..((((.......)))).....))).)))))))(((((....))))). (-31.20 = -31.77 + 0.56)


| Location | 15,368,011 – 15,368,131 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -35.29 |
| Energy contribution | -35.91 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15368011 120 - 22224390 CUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGAAGAGACACGCAAAGUGGCCUAUUAAAAUGCGCACAUAUAGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCAC ((((((((((..(((......)))))))..))))))...(((.....)))(((.((((...(((.(((((...((................))...)))))...)))....))))))).. ( -30.79) >DroSec_CAF1 825 120 - 1 CUUCCAGCGACAAGUGGUAAAGCUUCUCAGUGGCAGAGGCACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCAC ((..((((((.((((......)))).))..((((((.(((((.....)))))))))))...(((.(((((.((((................)))).)))))...)))..))))..))... ( -40.39) >DroSim_CAF1 3330 120 - 1 CUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGCAGAGGAACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCAC .(((((.(.....))))))..(((...(((((((((.((.((.....)).)))))))....(((.(((((.((((................)))).)))))...)))..))))..))).. ( -35.99) >DroEre_CAF1 7965 120 - 1 CUUCCUGCGACAAGUGGAAGCGCCACGCAGUGGCAGAGACACGCAAAGUGUCCUGUCAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAGAGCUGAUGGCAC (((((.((.....))))))).((((((((.((((((.(((((.....)))))))))))...))))(((((.((((................)))).))))).((((...)))).)))).. ( -48.29) >consensus CUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGCAGAGACACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUAAAUACGCAUACGCCAGGUGUGACAGCAAAGCUGAUGGCAC .((((.((.....))))))..(((...(((((((((.(((((.....))))))))))....(((.(((((.((((................)))).)))))...)))..))))..))).. (-35.29 = -35.91 + 0.62)


| Location | 15,368,051 – 15,368,171 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -28.67 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15368051 120 - 22224390 GUCCUUUUGGCGACAAUUAAGCCCCAUAAUGAGCUAUAAACUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGAAGAGACACGCAAAGUGGCCUAUUAAAAUGCGCACAUAUAGCAAAUA ........(((.........))).........((((((..((((((((((..(((......)))))))..))))))...........(((((..........)).))).))))))..... ( -27.30) >DroSec_CAF1 865 120 - 1 GUCCUUUUGGCGACAAUUAAGCCCCAUAAUGAGCUAUAAACUUCCAGCGACAAGUGGUAAAGCUUCUCAGUGGCAGAGGCACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUA (.((...(((((.((...((((.((((.....(((..........))).....))))....)))).....((((((.(((((.....)))))))))))...))))).))...)))..... ( -31.10) >DroSim_CAF1 3370 120 - 1 GUCCUUUUGGCGACAAUUAAGCCCCAUAAUGAGCUAUAAACUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGCAGAGGAACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUA ........(((.........)))(((((.((.((.......(((((.(.....)))))).......(((.((((((.((.((.....)).))))))))...))))).)))))))...... ( -29.50) >DroEre_CAF1 8005 120 - 1 GGCCUUUUGGCGACAAUUAAGCCCCAUAAUGAGCUAUAAACUUCCUGCGACAAGUGGAAGCGCCACGCAGUGGCAGAGACACGCAAAGUGUCCUGUCAAAAUGCGCACAUAUGGCAAAUA .(((...(((((.......(((.(......).))).....(((((.((.....))))))))))))((((.((((((.(((((.....)))))))))))...)))).......)))..... ( -42.70) >consensus GUCCUUUUGGCGACAAUUAAGCCCCAUAAUGAGCUAUAAACUUCCAGCGACAAGUGGAAAAGCUUCGCAGUGGCAGAGACACGCAAAGUGCCCUGUUAAAAUGCGCACAUAUGGCAAAUA ........(((.........))).........((((((...((((.((.....))))))......((((.((((((.(((((.....)))))))))))...))))....))))))..... (-28.67 = -29.30 + 0.63)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:44 2006