
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,367,435 – 15,367,698 |
| Length | 263 |
| Max. P | 0.991465 |

| Location | 15,367,435 – 15,367,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -33.05 |
| Energy contribution | -33.18 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367435 120 + 22224390 CACCCUCUUAUCAGCCCCAACCCGAGUUUAUUUUUAUCGUACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAA ((((.((..((((((..((...(((...........)))...))..))))))...))....(((((((((..(((.(((.....))))))....))))))))).....))))........ ( -35.40) >DroSec_CAF1 250 120 + 1 CACCCUCUUAUCAGACCCAACCCGAGUUUAUUUUUAUCGCACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAA ((((.((..(((((...((...(((...........)))...))...)))))...))....(((((((((..(((.(((.....))))))....))))))))).....))))........ ( -32.30) >DroSim_CAF1 2750 120 + 1 CACCCUCUUAUCAGCCCCAACCCGAGUUUAUUUUUAUCGCACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAUAGGUGAAAUGCAA ((((.((..((((((..((...(((...........)))...))..))))))...))....(((((((((..(((.(((.....))))))....))))))))).....))))........ ( -35.40) >DroEre_CAF1 7368 117 + 1 CGCCCUCUUAUCAGCCCCAACCCGAGUUUAUUUUUAUCGCACUG---CUGAUUAUGAAAUUCCUUUGGCAAAGGCUCUGUGAGGCAGGCCAAAGUGCCAAAGGUUAAAGGUGAAAGGUAA ((((.((..((((((.......(((...........)))....)---)))))...))....(((((((((..(((.(((.....))))))....))))))))).....))))........ ( -35.40) >consensus CACCCUCUUAUCAGCCCCAACCCGAGUUUAUUUUUAUCGCACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAA ((((.((..((((((..((...(((...........)))...))..))))))...))....(((((((((..(((.(((.....))))))....))))))))).....))))........ (-33.05 = -33.18 + 0.13)

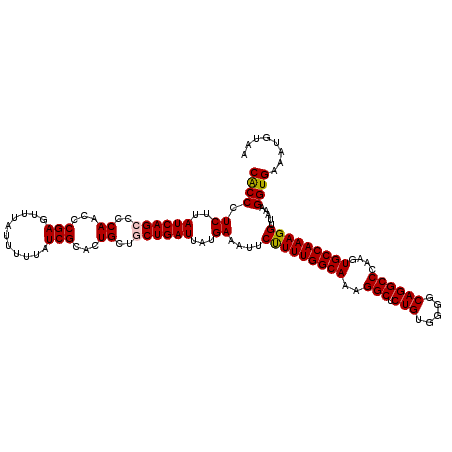
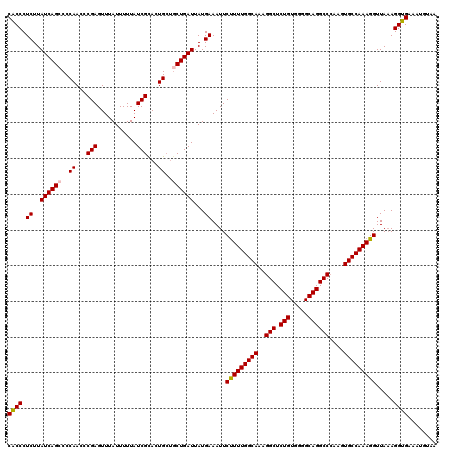
| Location | 15,367,435 – 15,367,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -31.25 |
| Energy contribution | -32.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367435 120 - 22224390 UUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGUACGAUAAAAAUAAACUCGGGUUGGGGCUGAUAAGAGGGUG ........(((((((..(.(((((((...(((((((.....))).)))).))))))).).........((((((..(((..(((...........)))..)))..))))))..))))))) ( -38.70) >DroSec_CAF1 250 120 - 1 UUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGUGCGAUAAAAAUAAACUCGGGUUGGGUCUGAUAAGAGGGUG ........(((((((..(.(((((((...(((((((.....))).)))).))))))).).........((((((..((((.(((...........))).)))).).)))))..))))))) ( -33.30) >DroSim_CAF1 2750 120 - 1 UUGCAUUUCACCUAUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGUGCGAUAAAAAUAAACUCGGGUUGGGGCUGAUAAGAGGGUG ........(((((....(.(((((((...(((((((.....))).)))).))))))).).........((((((..((((.(((...........))).))))..))))))....))))) ( -36.00) >DroEre_CAF1 7368 117 - 1 UUACCUUUCACCUUUAACCUUUGGCACUUUGGCCUGCCUCACAGAGCCUUUGCCAAAGGAAUUUCAUAAUCAG---CAGUGCGAUAAAAAUAAACUCGGGUUGGGGCUGAUAAGAGGGCG ..........(((((..(((((((((....((((((.....))).)))..))))))))).........(((((---(..(.(((...........))).).....)))))).)))))... ( -35.40) >consensus UUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGUGCGAUAAAAAUAAACUCGGGUUGGGGCUGAUAAGAGGGUG ........(((((((..(.(((((((...(((((((.....))).)))).))))))).).........((((((..(((..(((...........)))..)))..))))))..))))))) (-31.25 = -32.50 + 1.25)

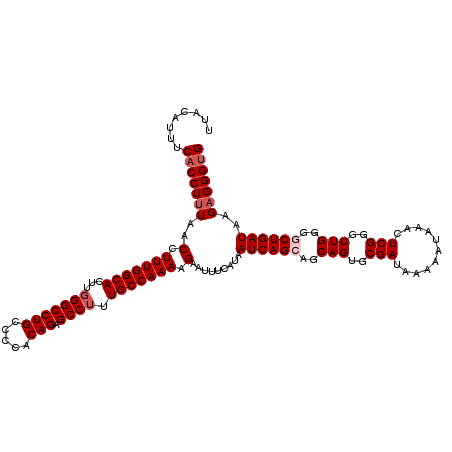

| Location | 15,367,475 – 15,367,578 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.06 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367475 103 + 22224390 ACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAAA----AGGUAAUAGGGGAUGGAGCUGU .(((.((((..((((......((((((((...(((.((...(((....))).)).)))....))))))))......)))).----.)))).)))............. ( -26.20) >DroSec_CAF1 290 91 + 1 ACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAAA----AGGCAACAGG------------ .(((.((((..((((......((((((((...(((.((...(((....))).)).)))....))))))))......)))).----.)))).))).------------ ( -29.90) >DroSim_CAF1 2790 96 + 1 ACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAUAGGUGAAAUGCAAAACAAAUGCAACAGGG----------- .(((.(((...((((((....(((((((((..(((.(((.....))))))....)))))))))))))))((.....)).........))).)))..----------- ( -27.90) >consensus ACUGCUGCUGAUUAUGAAAUUCUUUUGGCAAAGGCUCUGUGGGGCAGGCCCAAGUGCCAAAGGUUAAAGGUGAAAUGUAAA____AGGCAACAGGG___________ .(((.((((..((((......(((((((((..(((.(((.....))))))....)))))))))......)))).............)))).)))............. (-25.17 = -25.06 + -0.11)

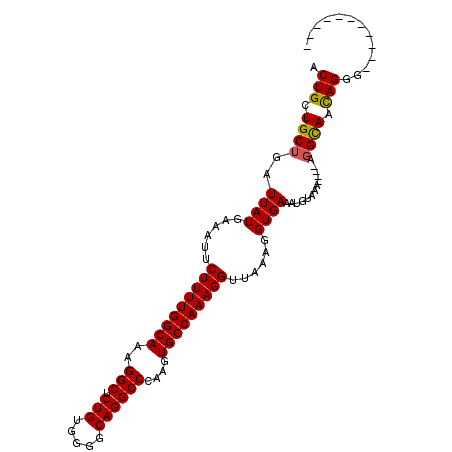
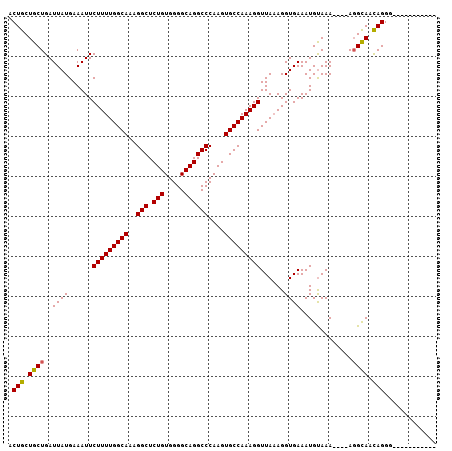
| Location | 15,367,475 – 15,367,578 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -22.47 |
| Energy contribution | -23.13 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367475 103 - 22224390 ACAGCUCCAUCCCCUAUUACCU----UUUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGU ...(((.(..............----....................(((((((...(((((((.....))).)))).))))))).(.....).......).)))... ( -19.70) >DroSec_CAF1 290 91 - 1 ------------CCUGUUGCCU----UUUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGU ------------.(((((((..----....................(((((((...(((((((.....))).)))).))))))).(.....).......))))))). ( -26.20) >DroSim_CAF1 2790 96 - 1 -----------CCCUGUUGCAUUUGUUUUGCAUUUCACCUAUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGU -----------..(((((((...((.....))............(.(((((((...(((((((.....))).)))).))))))).).............))))))). ( -27.80) >consensus ___________CCCUGUUGCCU____UUUACAUUUCACCUUUAACCUUUGGCACUUGGGCCUGCCCCACAGAGCCUUUGCCAAAAGAAUUUCAUAAUCAGCAGCAGU .............(((((((..........................(((((((...(((((((.....))).)))).))))))).(.....).......))))))). (-22.47 = -23.13 + 0.67)


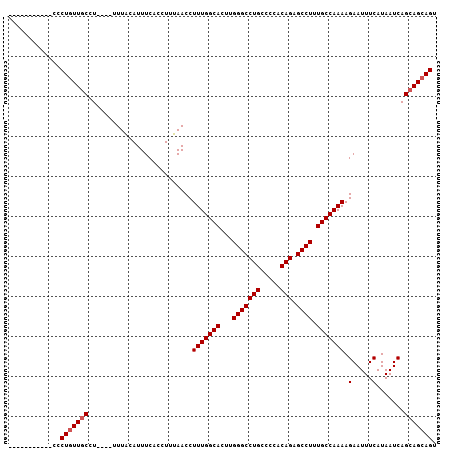
| Location | 15,367,555 – 15,367,658 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367555 103 - 22224390 UUUGCUGUGGUGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUUGCAUUGUUGGUCUGUCACCCCGCACCGCUUCACAGCUCCAUCCCCUAUUACCU----U ...((((((((((((......)))...........(((((((((((........).))))))))))........)))..)))))).................----. ( -24.70) >DroSec_CAF1 370 86 - 1 UUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC-----------------CCUGUUGCCU----U ..(((.(((((((.(((((...(((..(....((.....))....)..)))...))))).)))))))...)))..-----------------..........----. ( -21.80) >DroSim_CAF1 2870 91 - 1 UUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC----------------CCCUGUUGCAUUUGUU ..(((.(((((((.(((((...(((..(....((.....))....)..)))...))))).)))))))...)))..----------------................ ( -21.80) >consensus UUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC________________CCCUGUUGCCU____U ..((((((.(.((((......)))).).)))....(((((((((((........).))))))))))....))).................................. (-20.10 = -20.10 + -0.00)

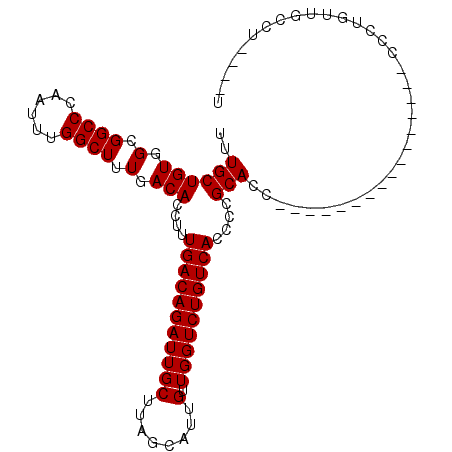
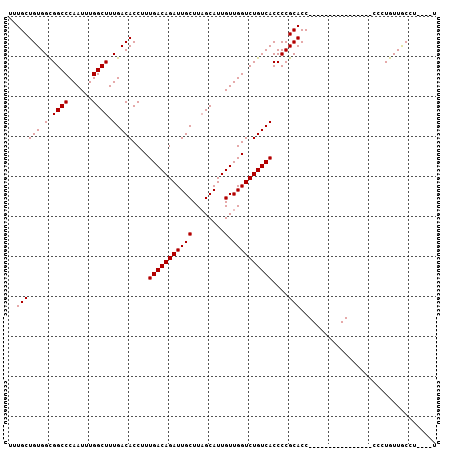
| Location | 15,367,578 – 15,367,698 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -29.31 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15367578 120 - 22224390 UUGUAAAGCAAACAAUUGUCGAAAAAUUAUUGCUGCCUCAUUUGCUGUGGUGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUUGCAUUGUUGGUCUGUCACCCCGCACCGCUUC .((((((((((....(((((((((((((...((..((.((.....)).))..))..)))))((........)))))))))).)))))))))).....(((.((......)).)))..... ( -30.20) >DroSec_CAF1 381 115 - 1 UUGUAAAGCAAACAAUUGUCGAAAAAUUAUUGCUGCCUCAUUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC----- .......((.......(((((((........((((((.((.....)).))))))((.....)).)))))))....(((((((((((........).))))))))))....))...----- ( -30.90) >DroSim_CAF1 2886 115 - 1 UUGUAAAGCAAACAAUUGUCGAAAAAUUAUUGCUGCCUCAUUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC----- .......((.......(((((((........((((((.((.....)).))))))((.....)).)))))))....(((((((((((........).))))))))))....))...----- ( -30.90) >DroEre_CAF1 7521 117 - 1 UUGUAAAGCAAACAAUUGUCGAAAAAUUAUUGCUGCCUCAUUUGCUGUGGCGGCCC-AUUUGGCUUUGACACCUUUGACAGAUUGCUUUGCAUUGUUGGUCUGUCACCCCUCACCC--UC .((((((((((....((((((((........((((((.((.....)).))))))((-....))..........)))))))).)))))))))).....(((.....)))........--.. ( -30.70) >consensus UUGUAAAGCAAACAAUUGUCGAAAAAUUAUUGCUGCCUCAUUUGCUGUGGCGGCCCAAUUUGGCUUUGACACCUUUGACAGAUUGCUUAGCAUUGUUGGUCUGUCACCCCGCACC_____ .((((((((((....((((((((........((((((.((.....)).)))))).......((........)))))))))).)))))))))).(((.((........)).)))....... (-29.31 = -29.88 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:39 2006