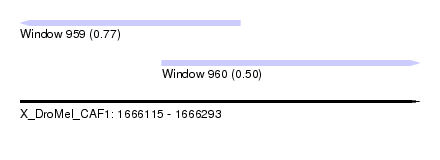
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,666,115 – 1,666,293 |
| Length | 178 |
| Max. P | 0.765363 |

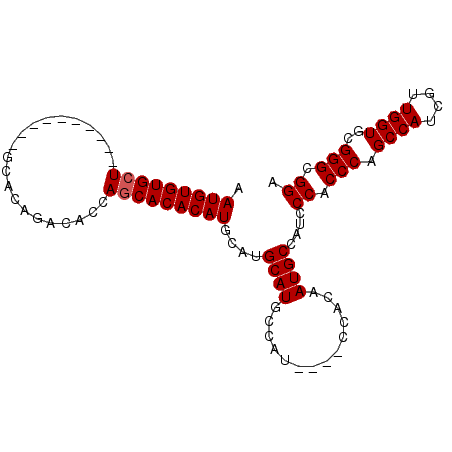
| Location | 1,666,115 – 1,666,213 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -24.37 |
| Energy contribution | -24.71 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1666115 98 - 22224390 AAUGUGUGCUGGCUGCAGCAGCACAGACACCAACACACAUGCAUGCAUGCCAUCCAUCCACAAUGCCAUCCCACCCAGCCAUCGUUGGUGCGGGCGGA ..((((((.(((((((((((...................))).)))).))))..))..))))........((.(((.((((....))))..))).)). ( -29.21) >DroSec_CAF1 449 84 - 1 AAUGUGUGCU----------GCACAGACCCCAGCACACAUGCAUGCAUGCCAU----CCACAAUGCCAUCCCACCCAGCCAUCGUUGGUGCGGGCGGA .(((((((((----------(.........))))))))))....((((.....----.....))))....((.(((.((((....))))..))).)). ( -30.20) >DroSim_CAF1 1329 84 - 1 AAUGUGUGCU----------GCACAGACACCAGCACACAUGCAUGCAUGCCAU----CCACAAUGCCAUCCCACCCAGCCAUCGUUGGUGCGGGCGGA .(((((((((----------(.........))))))))))....((((.....----.....))))....((.(((.((((....))))..))).)). ( -30.20) >consensus AAUGUGUGCU__________GCACAGACACCAGCACACAUGCAUGCAUGCCAU____CCACAAUGCCAUCCCACCCAGCCAUCGUUGGUGCGGGCGGA .(((((((((.....................)))))))))....((((..............))))....((.(((.((((....))))..))).)). (-24.37 = -24.71 + 0.33)

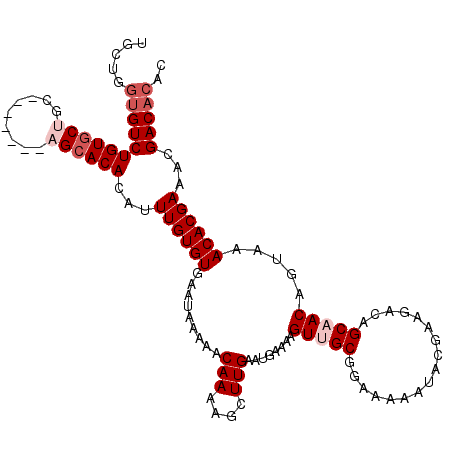
| Location | 1,666,178 – 1,666,293 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -17.75 |
| Energy contribution | -19.15 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1666178 115 + 22224390 UGUUGGUGUCUGUGCUGCUGCAGCCAGCACACAUUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCAACAGUAAACACGAAACGACACAC .....(((((((((((((....).)))))))..(((((((.........(((....)))........(((((.................))))).....)))))))..))))).. ( -29.03) >DroSec_CAF1 508 105 + 1 UGCUGGGGUCUGUGC----------AGCACACAUUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCAACAGUAAACACGAAACGACACAC (((((..(.((((((----------..(((((....)))))............(((((.......)))))))(........)....)))))..)))))................. ( -20.80) >DroSim_CAF1 1388 105 + 1 UGCUGGUGUCUGUGC----------AGCACACAUUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCAACAGUAAACACGAAACGACACAC .....(((((((((.----------....))))(((((((.........(((....)))........(((((.................))))).....)))))))..))))).. ( -21.33) >DroEre_CAF1 302 108 + 1 UGCUGCUGUCUGUGCUGC-------AGCACACACUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCUACAGUAAACACGAAACGACACAC (((((((((((...((((-------(((.((((....))))........(((....)))........)))))))..........)))))))...))))................. ( -25.02) >DroYak_CAF1 1447 108 + 1 AGCUGGUGUCUGUGCUGC-------AGAACACACUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCUACAGUAAACACGAAACGACACAC (((((...(((((.((((-------....((((....))))............(((((.......)))))))))....))).))...)))))....................... ( -20.70) >consensus UGCUGGUGUCUGUGCUGC_______AGCACACAUUUGUGUGAAUAAAAACAAAAGCUUGAAUGAAAAGUUGCGGAAAAAUACGAAGACAGCAACAGUAAACACGAAACGACACAC .....(((((((((((.........))))))...((((((.........(((....)))........(((((.................))))).....))))))...))))).. (-17.75 = -19.15 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:57 2006