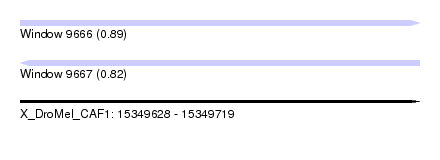
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,349,628 – 15,349,719 |
| Length | 91 |
| Max. P | 0.894349 |

| Location | 15,349,628 – 15,349,719 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.37 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
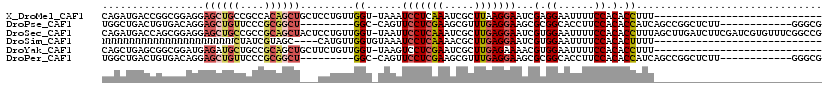
>X_DroMel_CAF1 15349628 91 + 22224390 CAGAUGACCGGCGGAGGAGCUGCCGCCACAGCUGCUCCUGUUGGU-UAAAUCCUCAAAUCGCUUAAGGAAUCGAGGAAUUUUCCACACCUUU---------------------------- ....((((((((((((.(((((......))))).)))).))))))-))..(((((...((.......))...)))))...............---------------------------- ( -31.80) >DroPse_CAF1 12487 98 + 1 UGGCUGACUGUGACAGGAGCUGUUCCCGCGGCU---------GGC-CAGUUCCUCGAAGCGUUUGAGGAAGCGCGGCACCUUCCACACCAUCAGCCGGCUCUU------------GGGCG .((((((.((((..(((((((((....))))))---------.((-(.(((((((((.....)))))).)))..))).)))..))))...)))))).(((...------------.))). ( -42.50) >DroSec_CAF1 8586 119 + 1 CAGAUGACCAGCGGAGGAGCUGCCGCCGCAGCUACUCCUGUUGGU-UAAUUCCUCAAAUCGCUUGAGGAAUCGUGGAAUUUUCCACACCUUUAGCUUGAUCUUCGAUCGUGUUUCGGCCG ....((((((((((((.((((((....)))))).)))).))))))-))(((((((((.....))))))))).(((((....))))).((...(((.(((((...))))).)))..))... ( -49.50) >DroSim_CAF1 8403 88 + 1 NNNNNNNNNNNNNNNNNNNNNNCUAUCGUAGC----CAUGUUGGUGUAAAUCCUCAAAACGCUUGAGGAAUCGUGGAAUUUUCCACACUUUU---------------------------- ..............................((----(.....))).....(((((((.....)))))))...(((((....)))))......---------------------------- ( -17.90) >DroYak_CAF1 10004 91 + 1 CAGCUGAGCGGCGGAUGAGAUGCUGCCGCAGCUGCUUCUGUUGGU-UAAGUCCUCGAAUCGCUUGAGAAAACGUGGAAUUUUCCACACCUUU---------------------------- (((((..(((((((........))))))))))))........(((-......(((((.....))))).....(((((....))))))))...---------------------------- ( -30.70) >DroPer_CAF1 13152 98 + 1 UGGCUGACUGUGACAGGAGCUGUUCCCGCGGCU---------GGC-CAGUUCCUCGAAGCGUUUGAGGAAGCGCGGCACCUUCCACACCAUCAGCCGGCUCUU------------GGGCG .((((((.((((..(((((((((....))))))---------.((-(.(((((((((.....)))))).)))..))).)))..))))...)))))).(((...------------.))). ( -42.50) >consensus CAGCUGACCGGCGGAGGAGCUGCUGCCGCAGCU___CCUGUUGGU_UAAUUCCUCAAAUCGCUUGAGGAAUCGUGGAAUUUUCCACACCUUU____________________________ .................((((((....)))))).........((......(((((((.....)))))))...(.((......)).).))............................... (-12.20 = -12.85 + 0.65)
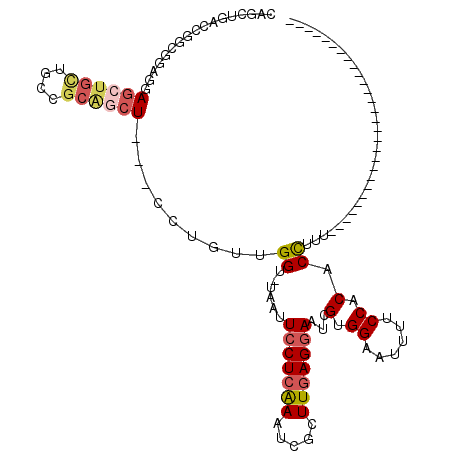

| Location | 15,349,628 – 15,349,719 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.37 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -12.22 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
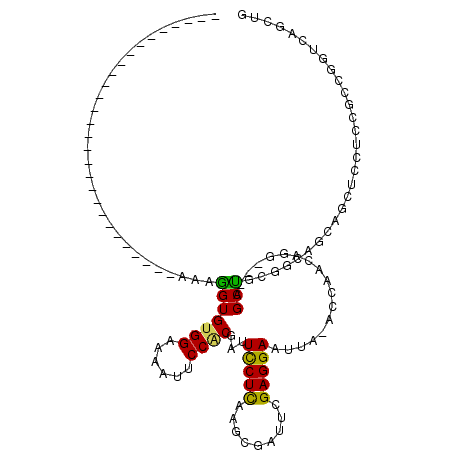
>X_DroMel_CAF1 15349628 91 - 22224390 ----------------------------AAAGGUGUGGAAAAUUCCUCGAUUCCUUAAGCGAUUUGAGGAUUUA-ACCAACAGGAGCAGCUGUGGCGGCAGCUCCUCCGCCGGUCAUCUG ----------------------------..((((((((..((.(((((((.(((....).)).))))))).)).-.)))...((((.((((((....)))))).))))......))))). ( -32.40) >DroPse_CAF1 12487 98 - 1 CGCCC------------AAGAGCCGGCUGAUGGUGUGGAAGGUGCCGCGCUUCCUCAAACGCUUCGAGGAACUG-GCC---------AGCCGCGGGAACAGCUCCUGUCACAGUCAGCCA .((..------------....)).(((((((..((((...(((((((.(.((((((.........)))))))))-)).---------.)))((((((.....))))))))))))))))). ( -38.10) >DroSec_CAF1 8586 119 - 1 CGGCCGAAACACGAUCGAAGAUCAAGCUAAAGGUGUGGAAAAUUCCACGAUUCCUCAAGCGAUUUGAGGAAUUA-ACCAACAGGAGUAGCUGCGGCGGCAGCUCCUCCGCUGGUCAUCUG .((((.......((((...)))).(((....((((((((....)))))(((((((((((...))))))))))).-)))....((((.((((((....)))))).)))))))))))..... ( -50.30) >DroSim_CAF1 8403 88 - 1 ----------------------------AAAAGUGUGGAAAAUUCCACGAUUCCUCAAGCGUUUUGAGGAUUUACACCAACAUG----GCUACGAUAGNNNNNNNNNNNNNNNNNNNNNN ----------------------------...((((((((....)))))((.((((((((...)))))))).))...........----)))............................. ( -15.70) >DroYak_CAF1 10004 91 - 1 ----------------------------AAAGGUGUGGAAAAUUCCACGUUUUCUCAAGCGAUUCGAGGACUUA-ACCAACAGAAGCAGCUGCGGCAGCAUCUCAUCCGCCGCUCAGCUG ----------------------------...((((((((....)))))((((((...........))))))...-)))........((((((((((............)))))..))))) ( -26.90) >DroPer_CAF1 13152 98 - 1 CGCCC------------AAGAGCCGGCUGAUGGUGUGGAAGGUGCCGCGCUUCCUCAAACGCUUCGAGGAACUG-GCC---------AGCCGCGGGAACAGCUCCUGUCACAGUCAGCCA .((..------------....)).(((((((..((((...(((((((.(.((((((.........)))))))))-)).---------.)))((((((.....))))))))))))))))). ( -38.10) >consensus ____________________________AAAGGUGUGGAAAAUUCCACGAUUCCUCAAGCGAUUCGAGGAAUUA_ACCAACAGG___AGCUGCGGCAGCAGCUCCUCCGCCGGUCAGCUG ...............................(((((((......))))...(((((.........)))))..................)))............................. (-12.22 = -11.58 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:29 2006