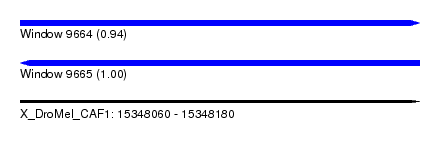
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,348,060 – 15,348,180 |
| Length | 120 |
| Max. P | 0.997951 |

| Location | 15,348,060 – 15,348,180 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -37.36 |
| Energy contribution | -38.28 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15348060 120 + 22224390 UCAAACGCCUCCAGUUGCUCCAAGUCCAGGAGCAGAAAGUUGGCGAUGAUCAACAGCACUGGCAUGGCCAUCAGAACGGCCAGGCAAAUGUGCCACGGAUACCAGCUGGAUAUGGCCUCG ......((((((..(((((((.......)))))))..((((((.....(((....((((..((.(((((........))))).))....))))....))).)))))))))...))).... ( -43.50) >DroPse_CAF1 10766 120 + 1 UCGAAGGCCUCCAGCUGCUCCAGGUCCAGCAGGAGGAAGUUAGCGAUGAUCAGGAGCACGGGCAUGGCCAUCAGCACGGCCAGGCAAAUAUGCCACGGAUACCAGCUGGAGAUGGCCUCG .(((.((((((((((((((((.((((..((............))...)))).)))))..((...(((((........)))))((((....)))).......)))))))))...))))))) ( -51.40) >DroSec_CAF1 7015 120 + 1 UCAAACGCCUCCAGUUGCUCCAAGUCCAGGAGCAGGAAGUUGGCGAUGAUCAACAGCACUGGCAUGGCCAUCAGAACGGCCAGGCAAAUGUGCCACGGAUACCAGCUGGAUAUGGCCUCG ......(((((((((((((((.......))))).((..(((((......))))).((((..((.(((((........))))).))....))))........)))))))))...))).... ( -44.80) >DroYak_CAF1 8439 120 + 1 UCAAACGCCUCCAGUUGCUCCAAGUCCAGGAGCAGAAAGUUGGCGAUGAUCAACAGCACGGGCAUGGCCAUCAGAACGGCCAGGCAAAUAUGCCACGGAUACCAGCUGGAUAUGGCCUCG ......(((((((((((((((.......))))).....((((..(.....)..))))..((...(((((........)))))((((....)))).......)))))))))...))).... ( -41.10) >DroPer_CAF1 11413 120 + 1 UCGAAGGCCUCCAGCUGCUCCAGGUCCAGCAGGAGGAAGUUGGCGAUGAUCAGGAGCACGGGCAUGGCCAUCAGCACGGCCAGGCAAAUGUGCCACGGAUACCAGCUGGAGAUGGCCUCG .(((.((((((((((((.(((..(.(((((........))))))........(..(((((.((.(((((........))))).))...)))))..))))...))))))))...))))))) ( -53.20) >consensus UCAAACGCCUCCAGUUGCUCCAAGUCCAGGAGCAGGAAGUUGGCGAUGAUCAACAGCACGGGCAUGGCCAUCAGAACGGCCAGGCAAAUGUGCCACGGAUACCAGCUGGAUAUGGCCUCG ......((((((((((((.((((.(((.......)))..)))).)...(((....(((((.((.(((((........))))).))...)))))....)))..))))))))...))).... (-37.36 = -38.28 + 0.92)

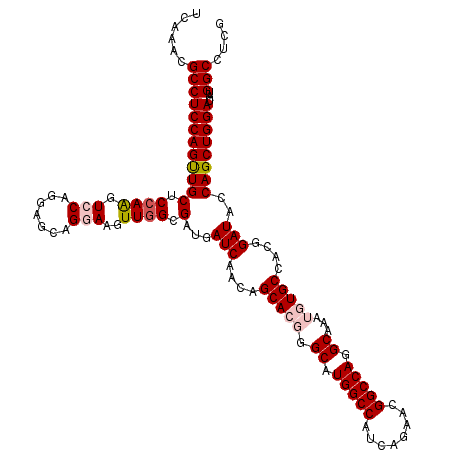
| Location | 15,348,060 – 15,348,180 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -49.10 |
| Consensus MFE | -42.72 |
| Energy contribution | -42.64 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15348060 120 - 22224390 CGAGGCCAUAUCCAGCUGGUAUCCGUGGCACAUUUGCCUGGCCGUUCUGAUGGCCAUGCCAGUGCUGUUGAUCAUCGCCAACUUUCUGCUCCUGGACUUGGAGCAACUGGAGGCGUUUGA (((((((...(((((.(((((((.(..((((....((.(((((((....))))))).))..))))..).)))....))))......((((((.......)))))).)))))))).)))). ( -47.70) >DroPse_CAF1 10766 120 - 1 CGAGGCCAUCUCCAGCUGGUAUCCGUGGCAUAUUUGCCUGGCCGUGCUGAUGGCCAUGCCCGUGCUCCUGAUCAUCGCUAACUUCCUCCUGCUGGACCUGGAGCAGCUGGAGGCCUUCGA (((((((...((((((((((((....((((....)))).((((((....)))))))))))...(((((.(.((...((............))..)).).)))))))))))))))).))). ( -52.20) >DroSec_CAF1 7015 120 - 1 CGAGGCCAUAUCCAGCUGGUAUCCGUGGCACAUUUGCCUGGCCGUUCUGAUGGCCAUGCCAGUGCUGUUGAUCAUCGCCAACUUCCUGCUCCUGGACUUGGAGCAACUGGAGGCGUUUGA (((((((...(((((.(((((((.(..((((....((.(((((((....))))))).))..))))..).)))....))))......((((((.......)))))).)))))))).)))). ( -47.70) >DroYak_CAF1 8439 120 - 1 CGAGGCCAUAUCCAGCUGGUAUCCGUGGCAUAUUUGCCUGGCCGUUCUGAUGGCCAUGCCCGUGCUGUUGAUCAUCGCCAACUUUCUGCUCCUGGACUUGGAGCAACUGGAGGCGUUUGA (((((((...(((((.(((((((.(..((((....((.(((((((....))))))).))..))))..).)))....))))......((((((.......)))))).)))))))).)))). ( -46.20) >DroPer_CAF1 11413 120 - 1 CGAGGCCAUCUCCAGCUGGUAUCCGUGGCACAUUUGCCUGGCCGUGCUGAUGGCCAUGCCCGUGCUCCUGAUCAUCGCCAACUUCCUCCUGCUGGACCUGGAGCAGCUGGAGGCCUUCGA (((((((...(((((((((.(((.(..((((....((.(((((((....))))))).))..))))..).))))....(((...(((.......)))..)))..)))))))))))).))). ( -51.70) >consensus CGAGGCCAUAUCCAGCUGGUAUCCGUGGCACAUUUGCCUGGCCGUUCUGAUGGCCAUGCCCGUGCUGUUGAUCAUCGCCAACUUCCUGCUCCUGGACUUGGAGCAACUGGAGGCGUUUGA (((((((...(((((.(((((((.(..((((....((.(((((((....))))))).))..))))..).)))....))))......((((((.......)))))).)))))))).)))). (-42.72 = -42.64 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:27 2006