
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,342,881 – 15,343,001 |
| Length | 120 |
| Max. P | 0.999339 |

| Location | 15,342,881 – 15,342,977 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.39 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
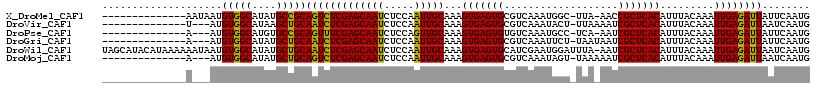
>X_DroMel_CAF1 15342881 96 + 22224390 ---------------------UACCAAUUGUAUCUGCAGCAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGGUU-UAA-GCCAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGA ---------------------......((((....))))..((((.(((((((...(((((((.(((((.((((-.((-....)).))).).))))))))))))...))))))).)))) ( -26.10) >DroVir_CAF1 25549 116 + 1 ACAGCCUCCUCUAUAUUGUUGUUAA--ACUUAUCUGCAGCAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUUUUAA-AGUAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGA ................((((((...--........))))))((((.(((((((...(((((((.(((((.(...((((-.....))))..).))))))))))))...))))))).)))) ( -28.30) >DroPse_CAF1 2149 95 + 1 ----------------------GCCGACUGUAUCUGCAGCAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUU-UGA-GGCAUUUGACACACUCACUUUGCAACUGGAGAUUGCUCGA ----------------------...(.((((....))))).((((.(((((((...(((((((.(((((.(..(-..(-(...))..)..).))))))))))))...))))))).)))) ( -25.40) >DroWil_CAF1 24251 100 + 1 U------UU------------UUUUUUUUGUAUCUGUAACAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUU-UAAAUCCAUUCGAUGCACUCACUUUGCAAUUGGAGAUUGCUCGA .------..------------....................((((.(((((((...(((((((.(((((.(...-...(((.....))).).))))))))))))...))))))).)))) ( -23.60) >DroMoj_CAF1 28643 100 + 1 A------------------UGUCCAACUCUUAUCUGCAGCAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUUUUUA-ACUAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGA .------------------......................((((.(((((((...(((((((.(((((.(....(((-(....))))..).))))))))))))...))))))).)))) ( -23.20) >DroAna_CAF1 1946 92 + 1 -------------------------AAAUCGAUCUGCAGCAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUU-UGA-GCCAUUUGACGGACUCACUUUGCAACUGGAGAUUGCUCGA -------------------------....((((((.(((.(((((......)))))............((((..-(((-(((.......)).))))..))))..))).))))))..... ( -23.50) >consensus _____________________UUCAAAUUGUAUCUGCAGCAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUU_UAA_GCCAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGA .........................................((((.(((((((...(((((((.(((((.(...................).))))))))))))...))))))).)))) (-21.23 = -21.39 + 0.17)

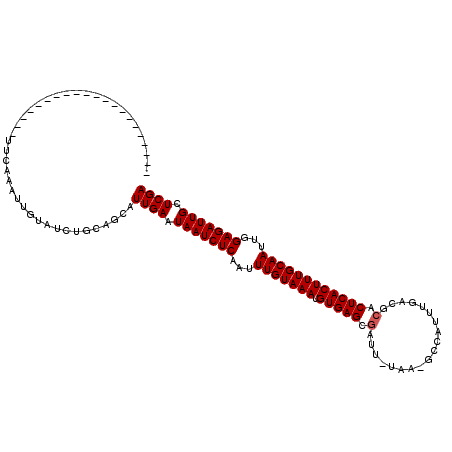

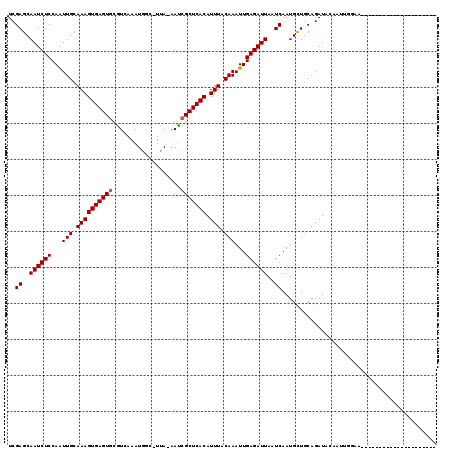
| Location | 15,342,881 – 15,342,977 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15342881 96 - 22224390 UCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUGGC-UUA-AACCGCUCACAUUUACAAAUUGAGAUUAUUCAAUGCUGCAGAUACAAUUGGUA--------------------- ..((....)).(((((((....(((((((.(((....)))-...-...)))))))((((.((.((((((....))))))..)).)))).)))))))..--------------------- ( -22.10) >DroVir_CAF1 25549 116 - 1 UCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUACU-UUAAAAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUGCUGCAGAUAAGU--UUAACAACAAUAUAGAGGAGGCUGU ....(((..((((..(((((..(((((((...........-.......)))))))........(((((......)))))..)))))....((--(....)))........))))..))) ( -25.07) >DroPse_CAF1 2149 95 - 1 UCGAGCAAUCUCCAGUUGCAAAGUGAGUGUGUCAAAUGCC-UCA-AAUCGCUCACAUUUACAAAUUGAGAUUAUUCAAUGCUGCAGAUACAGUCGGC---------------------- ..(((.((((((...(((.((((((((((...........-...-...))))))).))).)))...)))))).)))...((((......))))....---------------------- ( -21.93) >DroWil_CAF1 24251 100 - 1 UCGAGCAAUCUCCAAUUGCAAAGUGAGUGCAUCGAAUGGAUUUA-AAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUGUUACAGAUACAAAAAAAA------------AA------A ..((..((((((...(((.((((((((((.((..((.....)).-.))))))))).))).)))...))))))..))......................------------..------. ( -18.60) >DroMoj_CAF1 28643 100 - 1 UCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUAGU-UAAAAAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUGCUGCAGAUAAGAGUUGGACA------------------U ..((....))(((((((.....(((((((...........-.......)))))))((((.((.(((((......)))))..)).))))...)))))))..------------------. ( -20.27) >DroAna_CAF1 1946 92 - 1 UCGAGCAAUCUCCAGUUGCAAAGUGAGUCCGUCAAAUGGC-UCA-AAUCGCUCACAUUUACAAAUUGAGAUUAUUCAAUGCUGCAGAUCGAUUU------------------------- ..(((.((((((...(((.(((((((((((((...)))).-...-....)))))).))).)))...)))))).)))..................------------------------- ( -18.60) >consensus UCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUGGC_UUA_AAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUGCUGCAGAUACAAUUGGAA_____________________ ..((..((((((...(((.((((((((((...................))))))).))).)))...))))))..))........................................... (-17.29 = -17.46 + 0.17)


| Location | 15,342,899 – 15,343,001 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.28 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15342899 102 + 22224390 CAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGGUU-UAA-GCCAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGAGACUGCGGCAUAUGCCACAUUAUU-------------- ..((((.(((((((...(((((((.(((((.((((-.((-....)).))).).))))))))))))...))))))).))))...((.(((....))).)).....-------------- ( -32.10) >DroVir_CAF1 25586 100 + 1 CAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUUUUAA-AGUAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGAGAUUGCAGCUUAUGCCACAU---A-------------- ..((((.(((((((...(((((((.(((((.(...((((-.....))))..).))))))))))))...))))))).))))....(((.....))).....---.-------------- ( -26.60) >DroPse_CAF1 2166 99 + 1 CAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUU-UGA-GGCAUUUGACACACUCACUUUGCAACUGGAGAUUGCUCGAAACUGCGGCACAUGCCACAU---U-------------- ..((((.(((((((...(((((((.(((((.(..(-..(-(...))..)..).))))))))))))...))))))).))))...((.(((....))).)).---.-------------- ( -28.60) >DroGri_CAF1 24861 100 + 1 CAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUUAUUA-AGAAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGAGAUUGCAGCAUAUGCCACAU---U-------------- ..((((.(((((((...(((((((.(((((((.(((...-......))))))..)))))))))))...))))))).))))....(((.....))).....---.-------------- ( -26.00) >DroWil_CAF1 24272 117 + 1 CAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUU-UAAAUCCAUUCGAUGCACUCACUUUGCAAUUGGAGAUUGCUCGAGAUUGCAGCAUAUGCCACAUUAUUUUUUAUGUAUGCUA ..((((.(((((((...(((((((.(((((.(...-...(((.....))).).))))))))))))...))))))).))))......(((((((.................))))))). ( -29.93) >DroMoj_CAF1 28664 100 + 1 CAUUGAUUAAUCUCAAUUUGUAAAUGUGAGCGAUUUUUA-ACUAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGAGACUGCAGCAUAUGCCACAU---U-------------- ..((((.(((((((...(((((((.(((((.(....(((-(....))))..).))))))))))))...))))))).))))....(((.....))).....---.-------------- ( -25.80) >consensus CAUUGAAUAAUCUCAAUUUGUAAAUGUGAGCGAUU_UAA_ACCAUUUGACGCACUCACUUUGCAAUUGGAGAUUGCUCGAGACUGCAGCAUAUGCCACAU___U______________ ..((((.(((((((...(((((((.(((((.(...................).))))))))))))...))))))).))))....(((.....)))....................... (-24.50 = -24.28 + -0.22)

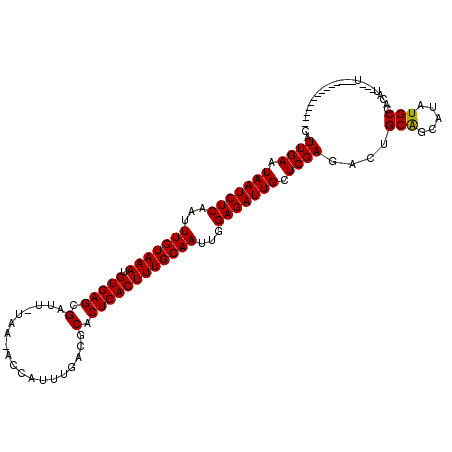
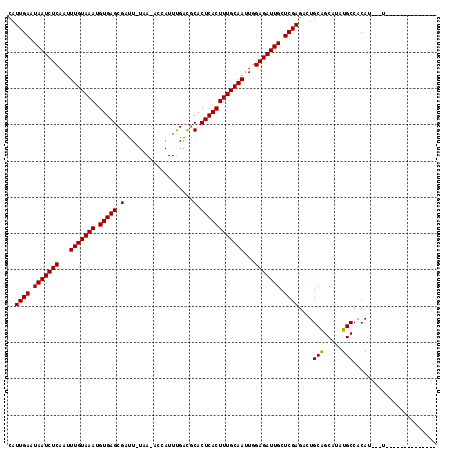
| Location | 15,342,899 – 15,343,001 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -27.09 |
| Energy contribution | -26.34 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15342899 102 - 22224390 --------------AAUAAUGUGGCAUAUGCCGCAGUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUGGC-UUA-AACCGCUCACAUUUACAAAUUGAGAUUAUUCAAUG --------------......(((((....)))))(((((((((((((.....)))))...(((((((.(((....)))-...-...))))))).........))))))))........ ( -30.70) >DroVir_CAF1 25586 100 - 1 --------------U---AUGUGGCAUAAGCUGCAAUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUACU-UUAAAAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUG --------------.---..(..((....))..)(((((((((((((.....)))))...(((((((...........-.......))))))).........))))))))........ ( -25.47) >DroPse_CAF1 2166 99 - 1 --------------A---AUGUGGCAUGUGCCGCAGUUUCGAGCAAUCUCCAGUUGCAAAGUGAGUGUGUCAAAUGCC-UCA-AAUCGCUCACAUUUACAAAUUGAGAUUAUUCAAUG --------------.---.((((((....)))))).....(((.((((((...(((.((((((((((...........-...-...))))))).))).)))...)))))).))).... ( -29.23) >DroGri_CAF1 24861 100 - 1 --------------A---AUGUGGCAUAUGCUGCAAUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUUCU-UAAUAAUCGCUCACAUUUACAAAUUGAGAUUAUUCAAUG --------------.---..(..((....))..)(((((((((((((.....)))))...(((((((...........-.......))))))).........))))))))........ ( -25.77) >DroWil_CAF1 24272 117 - 1 UAGCAUACAUAAAAAAUAAUGUGGCAUAUGCUGCAAUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCAUCGAAUGGAUUUA-AAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUG (((((((((((........))))...))))))).(((((((((((((.....)))))...(((((((.((..((.....)).-.))))))))).........))))))))........ ( -28.00) >DroMoj_CAF1 28664 100 - 1 --------------A---AUGUGGCAUAUGCUGCAGUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUAGU-UAAAAAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUG --------------.---..(..((....))..)(((((((((((((.....)))))...(((((((...........-.......))))))).........))))))))........ ( -25.77) >consensus ______________A___AUGUGGCAUAUGCUGCAAUCUCGAGCAAUCUCCAAUUGCAAAGUGAGUGCGUCAAAUGCU_UUA_AAUCGCUCACAUUUACAAAUUGAGAUUAAUCAAUG ....................(((((....)))))(((((((((((((.....)))))...(((((((...................))))))).........))))))))........ (-27.09 = -26.34 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:21 2006