
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,339,766 – 15,339,858 |
| Length | 92 |
| Max. P | 0.862302 |

| Location | 15,339,766 – 15,339,858 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.41 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -11.80 |
| Energy contribution | -13.43 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15339766 92 + 22224390 CAGCUGCUAGCCAAGGAGCAGCAA---CUUCAGCUAAACCAAGCUGAACUGGAAAAACUACAAGAGACACUUCGAGUUAACGAAGAGCAGCUCCU-------------------- ..((.....))..((((((.((..---.(((((((......))))))).....................(((((......))))).)).))))))-------------------- ( -26.50) >DroSim_CAF1 563 92 + 1 CAGCUGCUGGCCAAGGAGCAGCAA---CUCCAGCUAAACCAAGCUGAACUGGAAAAACUACAGGAGACACUGCGAGUUAACAAAGAGCAGCUCCU-------------------- .(((((((..(((.((((......---))))((((......))))....)))...((((.(((......)))..)))).......)))))))...-------------------- ( -26.70) >DroYak_CAF1 594 92 + 1 CAGCUGCUGGCCAAGGAGCAGCAA---AUGCAGCUAGACCAAGCUGAACUGGAAAAAUUACACGAGACACUUCGGGCAAAUGAAGAGCAAGUCCU-------------------- ..((((((........))))))..---...(((((......)))))....(((.........((((....)))).((.........))...))).-------------------- ( -21.30) >DroMoj_CAF1 24598 112 + 1 CUGCAGCUGGCCAAGCAGGAGCAGGAGGCGCAAUUGGGC---CUGCAGCUGGAGCAGCUUAGAGUGGAACUGCGUGCCAAGCAGGAGCAGCUGGAGCUGAAGUUACAACAAAAUG ((.((((((.((..((....)).))((((.(....).))---)).)))))).))((((((..(((.(..((((.......))))...).))).))))))................ ( -38.30) >consensus CAGCUGCUGGCCAAGGAGCAGCAA___CUGCAGCUAAACCAAGCUGAACUGGAAAAACUACAAGAGACACUGCGAGCUAACGAAGAGCAGCUCCU____________________ ..((((((..(((.(...((((....................))))..)))).................(((((......)))))))))))........................ (-11.80 = -13.43 + 1.62)

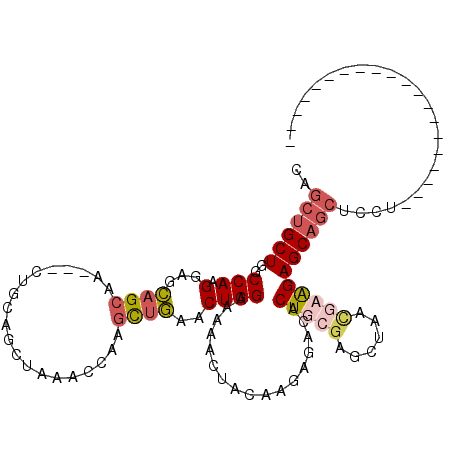

| Location | 15,339,766 – 15,339,858 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.41 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -14.56 |
| Energy contribution | -15.81 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
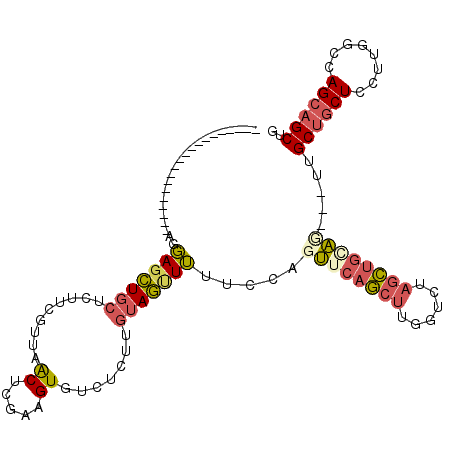
>X_DroMel_CAF1 15339766 92 - 22224390 --------------------AGGAGCUGCUCUUCGUUAACUCGAAGUGUCUCUUGUAGUUUUUCCAGUUCAGCUUGGUUUAGCUGAAG---UUGCUGCUCCUUGGCUAGCAGCUG --------------------.((((((((.(((((......)))))........)))))...)))...((((((......))))))((---((((((((....))).))))))). ( -30.90) >DroSim_CAF1 563 92 - 1 --------------------AGGAGCUGCUCUUUGUUAACUCGCAGUGUCUCCUGUAGUUUUUCCAGUUCAGCUUGGUUUAGCUGGAG---UUGCUGCUCCUUGGCCAGCAGCUG --------------------...(((((((....(((((...((((((.((((...((((...((((......))))...))))))))---.))))))...))))).))))))). ( -31.60) >DroYak_CAF1 594 92 - 1 --------------------AGGACUUGCUCUUCAUUUGCCCGAAGUGUCUCGUGUAAUUUUUCCAGUUCAGCUUGGUCUAGCUGCAU---UUGCUGCUCCUUGGCCAGCAGCUG --------------------.(((((..........((((.(((......))).)))).......)))))(((((((((.(((.((..---..)).)))....)))))..)))). ( -22.63) >DroMoj_CAF1 24598 112 - 1 CAUUUUGUUGUAACUUCAGCUCCAGCUGCUCCUGCUUGGCACGCAGUUCCACUCUAAGCUGCUCCAGCUGCAG---GCCCAAUUGCGCCUCCUGCUCCUGCUUGGCCAGCUGCAG ......((((......))))..((((((..(((((((((...((((((........)))))).))))..))))---).........(((....((....))..)))))))))... ( -33.80) >consensus ____________________AGGAGCUGCUCUUCGUUAACUCGAAGUGUCUCUUGUAGUUUUUCCAGUUCAGCUUGGUCUAGCUGCAG___UUGCUGCUCCUUGGCCAGCAGCUG ......................(((((((.........((.....)).......))))))).....((((((((......)))))))).....((((((........)))))).. (-14.56 = -15.81 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:14 2006