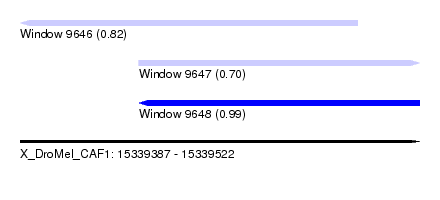
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,339,387 – 15,339,522 |
| Length | 135 |
| Max. P | 0.993694 |

| Location | 15,339,387 – 15,339,501 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -32.92 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15339387 114 - 22224390 UCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGAUUUUGGAGAUUGGCCAACUGGAGUUUGUGCUGAUCCUGACUGAUCUUCAUCUGAAUGGACUUC (((..(((.((((((((((...(((((.(((((.((((((.......))))))..)))))...((.(((....))).)))))))....)))))..))))).)))...))).... ( -33.70) >DroSim_CAF1 184 114 - 1 UCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGAUUUUGGAGAUUGGCCAACUGGAGUUUGUGCUGAUCCUGACUGUUCUUCAUCUGAAUGGACUUC (((..(((..(((((((((...(((((.(((((.((((((.......))))))..)))))...((.(((....))).)))))))....)))))))))....)))...))).... ( -35.20) >DroYak_CAF1 215 114 - 1 UCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAGCUGAUUUUGGAGAUUGGCUACCUGGAUUUUGUGCUGAUCCUGACUGAUCUUCAGCUGAACGGAUUUC (((..((((((((((((((....((((((((((.(((((((.....)))))))..)))))..)))))...(((((.......))))).)))))..)))))))))...))).... ( -46.90) >consensus UCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGAUUUUGGAGAUUGGCCAACUGGAGUUUGUGCUGAUCCUGACUGAUCUUCAUCUGAAUGGACUUC (((..(((.((((((((((...(((((.(((((.((((((.......))))))..)))))....((....)).......)))))....)))))..))))).)))...))).... (-32.92 = -32.70 + -0.22)



| Location | 15,339,427 – 15,339,522 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 60.89 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.32 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15339427 95 + 22224390 CUCCAGUUGGCCAAUCUCCAAAA------UCAGUUGCAAGCCGAUCAGGAGAAGCUAAGAGAACUGCUCCAGCUGCAGGACAAACUGGAGCAGCAAAAGGA---------- .(((..(((((...(((((...(------((.((.....)).)))..))))).))))).....(((((((((.(........).))))))))).....)))---------- ( -30.10) >DroVir_CAF1 20437 100 + 1 ---UGGCGAACUGAUC--GAGCGUGUGCGUCAGGUUGAUGCCGAGCGCGAGAAGCUG------CAACUUGAGUUGGAUGAGGUUGUCAAACAGGCGAAGGAAAAGAAAACU ---((((((.((.(((--.(((...(((((..(((....)))..)))))....))).------((((....))))))).)).))))))....................... ( -28.90) >DroSim_CAF1 224 95 + 1 CUCCAGUUGGCCAAUCUCCAAAA------UCAGUUGCAAGCCGAUCAGGAGAAGCUAAGAGAACUGCUCCAGCUGCAGGACAAACUGGAGCAGCAAAAGGA---------- .(((..(((((...(((((...(------((.((.....)).)))..))))).))))).....(((((((((.(........).))))))))).....)))---------- ( -30.10) >DroYak_CAF1 255 95 + 1 AUCCAGGUAGCCAAUCUCCAAAA------UCAGCUGCAAGCCGAUCAGGAGAAGCUAAGAGAACUGCUCCAGCUGCAGGACAAACUGGAGCAGCAAAAGGA---------- .(((...((((...(((((...(------((.((.....)).)))..))))).))))......(((((((((.(........).))))))))).....)))---------- ( -31.70) >DroMoj_CAF1 23391 100 + 1 ---CGGUGAGCUGAUC--GAGCGCGUCCGGCAAGCGGAUGCCGAGCGGGACAAAUUG------CAGCGCGAGCUGGAGGAGCUGGUGAAGCAGUCCAAGGAGAAGAAAACG ---..((...((..((--..(((((((((.....)))))))...)).((((...((.------((((....)))).))..(((.....))).))))...))..))...)). ( -35.70) >consensus _UCCAGUUAGCCAAUCUCCAAAA______UCAGGUGCAAGCCGAUCAGGAGAAGCUAAGAGAACUGCUCCAGCUGCAGGACAAACUGGAGCAGCAAAAGGA__________ ........................................((.....((((...............)))).(((.(((......))).))).......))........... ( -9.52 = -9.32 + -0.20)

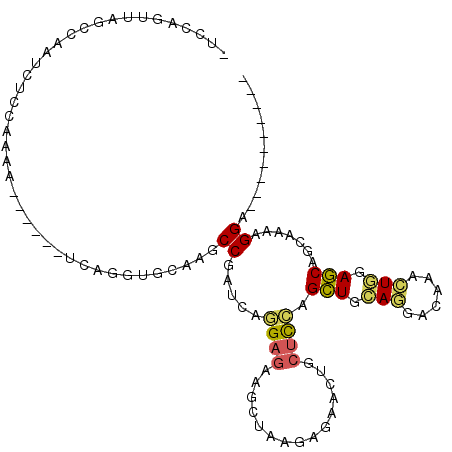

| Location | 15,339,427 – 15,339,522 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 60.89 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -13.67 |
| Energy contribution | -14.75 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15339427 95 - 22224390 ----------UCCUUUUGCUGCUCCAGUUUGUCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGA------UUUUGGAGAUUGGCCAACUGGAG ----------(((..((((((((((((((.(.....))))))))))))........((((((((.((((((.......))))------))..)))))..))))))..))). ( -37.00) >DroVir_CAF1 20437 100 - 1 AGUUUUCUUUUCCUUCGCCUGUUUGACAACCUCAUCCAACUCAAGUUG------CAGCUUCUCGCGCUCGGCAUCAACCUGACGCACACGCUC--GAUCAGUUCGCCA--- ...................((..(((.....)))..)).........(------(((((..(((.((...((.((.....)).))....)).)--))..)))).))..--- ( -14.50) >DroSim_CAF1 224 95 - 1 ----------UCCUUUUGCUGCUCCAGUUUGUCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGA------UUUUGGAGAUUGGCCAACUGGAG ----------(((..((((((((((((((.(.....))))))))))))........((((((((.((((((.......))))------))..)))))..))))))..))). ( -37.00) >DroYak_CAF1 255 95 - 1 ----------UCCUUUUGCUGCUCCAGUUUGUCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAGCUGA------UUUUGGAGAUUGGCUACCUGGAU ----------(((....((((((((((((.(.....))))))))))))).....((((((((((.(((((((.....)))))------))..)))))..)))))...))). ( -43.60) >DroMoj_CAF1 23391 100 - 1 CGUUUUCUUCUCCUUGGACUGCUUCACCAGCUCCUCCAGCUCGCGCUG------CAAUUUGUCCCGCUCGGCAUCCGCUUGCCGGACGCGCUC--GAUCAGCUCACCG--- .............(((((..(((.....)))...)))))...(.((((------....(((..(((..(((((......)))))..)).)..)--)).)))).)....--- ( -25.30) >consensus __________UCCUUUUGCUGCUCCAGUUUGUCCUGCAGCUGGAGCAGUUCUCUUAGCUUCUCCUGAUCGGCUUGCAACUGA______UUUUGGAGAUUGGCUAACUGGA_ .................((((((((((((........)))))))))))).......((((((((.((((((.......)))).......)).)))))..)))......... (-13.67 = -14.75 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:12 2006