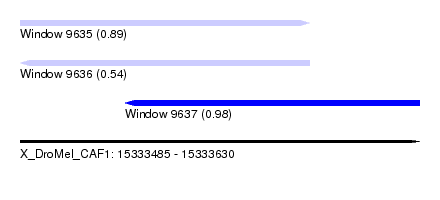
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,333,485 – 15,333,630 |
| Length | 145 |
| Max. P | 0.976941 |

| Location | 15,333,485 – 15,333,590 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -9.89 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15333485 105 + 22224390 --CAGAAAGAGACGGGGCUAGUCGUAGAGUGACUUGGCUUAAACU----UGAGCUAACUUGAGUUAUUC-CUCGCUU------UUUUAGCCCCAUCUCGC--UUUUUUUUUCGUUUAAGC --......((((.((((((((.((..(((((((((((((((....----)))))).....)))))))))-..))...------..)))))))).))))((--((............)))) ( -36.30) >DroSec_CAF1 38168 85 + 1 --CAGAAAGAGACGGGGCUAGUCGCAGAGUGACUUGGCUUAAACU----UGAGCUAAGUUGAGUUAUUC-CUCGCUU------UU----------------------UUUUCGUUUAAGC --.((((((((.((((((.....)).((((((((..(((((....----.....)))))..))))))))-)))))))------))----------------------))).......... ( -26.90) >DroSim_CAF1 14849 84 + 1 --CAGAAAGAGACGGGGCUAGUCGCAGAGUGACUUGGCUUAAACU----UGAGCUAAGUUGAGUUAUUC-CUCGCUU------UU-----------------------UUUCGUUUAAGC --..((((((((((((((.....)).((((((((..(((((....----.....)))))..))))))))-)))).))------))-----------------------))))........ ( -28.00) >DroEre_CAF1 13897 104 + 1 --GCGAAAGAGACGGGGCUAGUCGCAGAGUGACUUGGCUUAAACU----UGAGCUAAGUUGAGUUAUUC-CUCGCUU------UUUUAGCCCCAUCUCGC---UUUUUUUUCGUUUAAGC --((((((((((.((((((((....((((((((((((((((....----)))))))))))(((......-)))))))------).)))))))).))))..---.....))))))...... ( -39.41) >DroYak_CAF1 14380 107 + 1 --GAGAGGGAGACGGAGCUAGUCGUAGAGUGACUUGGCUUUAACU----UGAGCUAACUUGAGUUAUUC-CUCGCCU------UUUUAGCCACAUCUCGCGCUUUUUUUUUCGUUUAAGC --(((.(.((((.(..(((((.((..((((((((((((((.....----.))))).....)))))))))-..))...------..)))))..).)))).).)))................ ( -28.90) >DroAna_CAF1 36312 111 + 1 GGCAGAAAGAGCCAGCGAUAGUC----AGAGACUUGGCUAGAAAAUAACUCAUCUCACCUGUGCUGUUAGCCCGCUUUUAGUUUUUUUUCCCCAUUU-----UUUUUGCUGCUUUUUAGC .((..(((((((.((((((((((----((....)))))))(((((.................((((..((....))..))))...))))).......-----...)))))))))))).)) ( -21.55) >consensus __CAGAAAGAGACGGGGCUAGUCGCAGAGUGACUUGGCUUAAACU____UGAGCUAACUUGAGUUAUUC_CUCGCUU______UUUUAGCCCCAUCU______UUUUUUUUCGUUUAAGC .........((((((((...((((.....))))((((((((........))))))))...................................................)))))))).... ( -9.89 = -11.20 + 1.31)

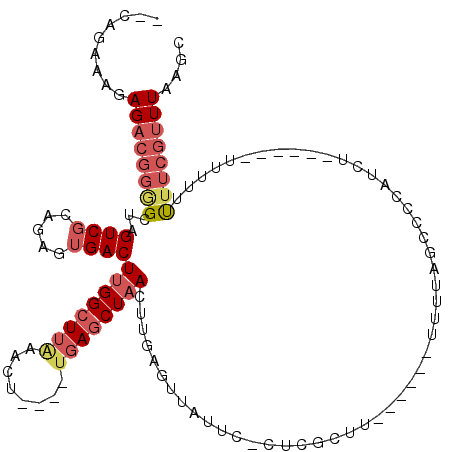

| Location | 15,333,485 – 15,333,590 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -8.49 |
| Energy contribution | -8.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15333485 105 - 22224390 GCUUAAACGAAAAAAAAA--GCGAGAUGGGGCUAAAA------AAGCGAG-GAAUAACUCAAGUUAGCUCA----AGUUUAAGCCAAGUCACUCUACGACUAGCCCCGUCUCUUUCUG-- ((((............))--))((((((((((((...------......(-(...((((..((....))..----))))....))..(((.......)))))))))))))))......-- ( -32.80) >DroSec_CAF1 38168 85 - 1 GCUUAAACGAAAA----------------------AA------AAGCGAG-GAAUAACUCAACUUAGCUCA----AGUUUAAGCCAAGUCACUCUGCGACUAGCCCCGUCUCUUUCUG-- ((((((((.....----------------------..------(((.(((-......)))..)))......----.))))))))..((((.......)))).................-- ( -15.04) >DroSim_CAF1 14849 84 - 1 GCUUAAACGAAA-----------------------AA------AAGCGAG-GAAUAACUCAACUUAGCUCA----AGUUUAAGCCAAGUCACUCUGCGACUAGCCCCGUCUCUUUCUG-- ((((((((....-----------------------..------(((.(((-......)))..)))......----.))))))))..((((.......)))).................-- ( -14.62) >DroEre_CAF1 13897 104 - 1 GCUUAAACGAAAAAAAA---GCGAGAUGGGGCUAAAA------AAGCGAG-GAAUAACUCAACUUAGCUCA----AGUUUAAGCCAAGUCACUCUGCGACUAGCCCCGUCUCUUUCGC-- ((((...........))---))((((((((((((...------..(((((-......))).((((.(((..----......))).))))......))...))))))))))))......-- ( -35.10) >DroYak_CAF1 14380 107 - 1 GCUUAAACGAAAAAAAAAGCGCGAGAUGUGGCUAAAA------AGGCGAG-GAAUAACUCAAGUUAGCUCA----AGUUAAAGCCAAGUCACUCUACGACUAGCUCCGUCUCCCUCUC-- ((((............))))..((((((..((((...------.((((((-......)))...(((((...----.))))).)))..(((.......)))))))..))))))......-- ( -29.80) >DroAna_CAF1 36312 111 - 1 GCUAAAAAGCAGCAAAAA-----AAAUGGGGAAAAAAAACUAAAAGCGGGCUAACAGCACAGGUGAGAUGAGUUAUUUUCUAGCCAAGUCUCU----GACUAUCGCUGGCUCUUUCUGCC ........((((.(((..-----...(((..........))).....((((...((((...(((.(((.((....)).))).))).((((...----))))...))))))))))))))). ( -21.80) >consensus GCUUAAACGAAAAAAAA______AGAUGGGGCUAAAA______AAGCGAG_GAAUAACUCAACUUAGCUCA____AGUUUAAGCCAAGUCACUCUGCGACUAGCCCCGUCUCUUUCUG__ ((((((((............((........))...............(((.......)))................))))))))..((((.......))))................... ( -8.49 = -8.93 + 0.45)

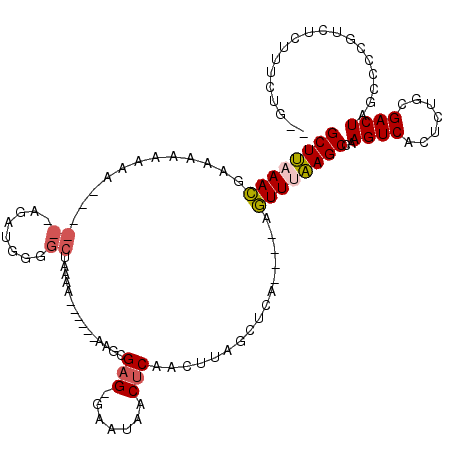

| Location | 15,333,523 – 15,333,630 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -18.84 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15333523 107 - 22224390 GGAGAUGUCAAGACGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAAAAAAAA--GCGAGAUGGGGCUAAAAAAGCGAGGAAUAACUCAAGUUAGCUCAAGUUU .(((.((((..(((.((((....))))...)))..))))..)))..............--........(((((((......(((......)))...)))))))...... ( -24.90) >DroSec_CAF1 38206 87 - 1 GGAGAUGUCAAGACGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAAA----------------------AAAAGCGAGGAAUAACUCAACUUAGCUCAAGUUU .(((.((((..(((.((((....))))...)))..)))).((((.........----------------------..))))(((......))).......)))...... ( -19.70) >DroSim_CAF1 14887 86 - 1 GGAGAUGUCAAGACGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAA-----------------------AAAAGCGAGGAAUAACUCAACUUAGCUCAAGUUU .(((.((((..(((.((((....))))...)))..)))).((((........-----------------------..))))(((......))).......)))...... ( -19.80) >DroEre_CAF1 13935 106 - 1 GGAGAUGUCAAGACGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAAAAAAA---GCGAGAUGGGGCUAAAAAAGCGAGGAAUAACUCAACUUAGCUCAAGUUU .(((.((((..(((.((((....))))...)))..))))..))).............---..(((.(((((((.....)))(((......)))..)))).)))...... ( -25.70) >DroYak_CAF1 14418 109 - 1 GGAGAUGUCAAGAUGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAAAAAAAAGCGCGAGAUGUGGCUAAAAAGGCGAGGAAUAACUCAAGUUAGCUCAAGUUA .(((..(((......((((....))))..(((((...((.((((............)))).)).....))))).....)))(((......))).......)))...... ( -24.80) >consensus GGAGAUGUCAAGACGCGACAAAAGUCGAAAGUCAAGGCGGGCUUAAACGAAAAAAAA___GCGAGAUG_GGCUAAAAAAGCGAGGAAUAACUCAACUUAGCUCAAGUUU ......(((..(((.((((....))))...)))..)))(((((.................((........)).........(((......))).....)))))...... (-18.84 = -18.88 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:03 2006