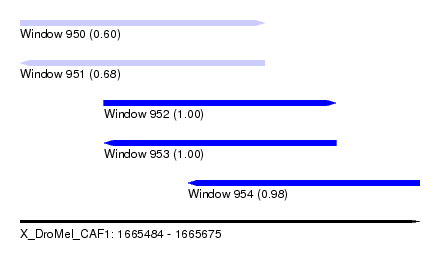
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,665,484 – 1,665,675 |
| Length | 191 |
| Max. P | 0.999960 |

| Location | 1,665,484 – 1,665,601 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -20.55 |
| Energy contribution | -23.00 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665484 117 + 22224390 CCUAAUUAGAUUACAUGCAAAUUAAAUGCAGCACGUAGAUACAUAUGUAUGGGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUGGUGGUGUCGAGCGU---UUGGGCUCAUG .............((((....((((((((.((((.(..((((....))))).))))(..(((((((.((((.((((....)))).)))))))))))..)...))))---))))...)))) ( -36.00) >DroSim_CAF1 683 114 + 1 CCUAAUUAGAUUACAUGCAAAUUAAAUGCAGCACGUACAUGCAUAUGUAUGUGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUG---GCGUUGAGCGU---UUGGGCUCAUU .........................(((((.(((((((((....))))))))))))))(((((((....((.((((....)))).)))))))---))..(((((..---....))))).. ( -37.60) >DroYak_CAF1 729 98 + 1 CCUAAUUAGAUUACAUGCAAAUUAAAUGCAGCACGU------------AUGUGUGCGUGCCACUAC-------UGUACAUGCUCGAUUGGUG---CUGACGAGCGAGCGUUGAGCUUAUG ...........((((((((.......))))((((((------------(....)))))))......-------))))...(((((((...((---((....))))...)))))))..... ( -26.50) >consensus CCUAAUUAGAUUACAUGCAAAUUAAAUGCAGCACGUA_AU_CAUAUGUAUGUGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUG___GUGUCGAGCGU___UUGGGCUCAUG .((((((........((((.......))))((((((((((....))))))))))((((((((((....)))))....)))))..))))))..........((((.........))))... (-20.55 = -23.00 + 2.45)

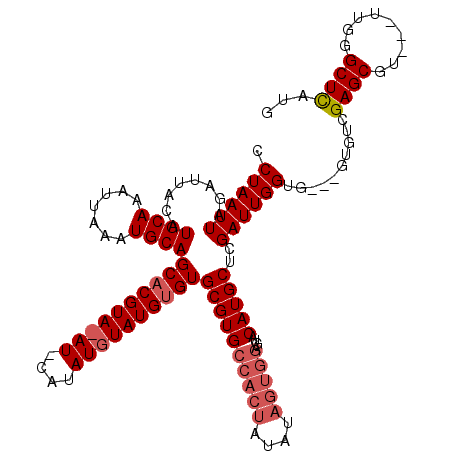
| Location | 1,665,484 – 1,665,601 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665484 117 - 22224390 CAUGAGCCCAA---ACGCUCGACACCACCACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACCCAUACAUAUGUAUCUACGUGCUGCAUUUAAUUUGCAUGUAAUCUAAUUAGG ..(((((....---..))))).................(((((((...((((....))))(((.((((..((((....))))....)))))))........)))))))............ ( -26.00) >DroSim_CAF1 683 114 - 1 AAUGAGCCCAA---ACGCUCAACGC---CACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACACAUACAUAUGCAUGUACGUGCUGCAUUUAAUUUGCAUGUAAUCUAAUUAGG ..(((((....---..)))))....---..........(((((((..(((((....)))))...((((((.(((((....))))).))).)))........)))))))............ ( -29.10) >DroYak_CAF1 729 98 - 1 CAUAAGCUCAACGCUCGCUCGUCAG---CACCAAUCGAGCAUGUACA-------GUAGUGGCACGCACACAU------------ACGUGCUGCAUUUAAUUUGCAUGUAAUCUAAUUAGG .....((.....((..(((((....---.......)))))..))...-------...(..(((((.......------------.)))))..).........))................ ( -21.20) >consensus CAUGAGCCCAA___ACGCUCGACAC___CACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACACAUACAUAUG_AU_UACGUGCUGCAUUUAAUUUGCAUGUAAUCUAAUUAGG ..(((((.........))))).................(((((((.........((.(..(((((....................)))))..)))......)))))))............ (-18.08 = -17.98 + -0.11)

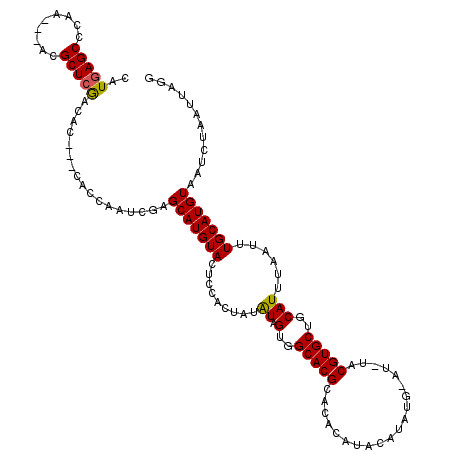
| Location | 1,665,524 – 1,665,635 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.13 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -23.71 |
| Energy contribution | -26.10 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665524 111 + 22224390 ACAUAUGUAUGGGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUGGUGGUGUCGAGCGU---UUGGGCUCAUGUACAAACA------UGAUGGUAAUGGCCGAUUUUUGGCCC .(((((((....((((((((((((((.((((.((((....)))).))))))))))))...((((..---....))))))))))..)))------).))).....((((((...)))))). ( -40.70) >DroSim_CAF1 723 114 + 1 GCAUAUGUAUGUGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUG---GCGUUGAGCGU---UUGGGCUCAUUUACAUACAUGUACAUGGUAGUAAUGGCCGAUUUUUGGCCC ((.((((((((((((.(((((((((....((.((((....)))).)))))))---))..(((((..---....)))))...)).))))))))))))))......((((((...)))))). ( -42.70) >DroYak_CAF1 765 89 + 1 --------AUGUGUGCGUGCCACUAC-------UGUACAUGCUCGAUUGGUG---CUGACGAGCGAGCGUUGAGCUUAUGUACA-------------UGGUAAUGGCCGAUUUUUGGCCA --------.........(((((....-------((((((((((((((...((---((....))))...)))))))..)))))))-------------))))).(((((((...))))))) ( -32.60) >consensus _CAUAUGUAUGUGUGCGUGCCACUAUAUAGUGGAGUACAUGCUCGAUUGGUG___GUGUCGAGCGU___UUGGGCUCAUGUACA_ACA______UG_UGGUAAUGGCCGAUUUUUGGCCC ...((((((((((.((((((..(........)..)))).((((((((..........))))))))........)).))))))))))..................((((((...)))))). (-23.71 = -26.10 + 2.39)

| Location | 1,665,524 – 1,665,635 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -15.55 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.52 |
| SVM decision value | 3.92 |
| SVM RNA-class probability | 0.999708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665524 111 - 22224390 GGGCCAAAAAUCGGCCAUUACCAUCA------UGUUUGUACAUGAGCCCAA---ACGCUCGACACCACCACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACCCAUACAUAUGU ((((........((......)).(((------(((....))))))))))..---..((((((............)))))).(((...(((((....)))))...)))............. ( -29.70) >DroSim_CAF1 723 114 - 1 GGGCCAAAAAUCGGCCAUUACUACCAUGUACAUGUAUGUAAAUGAGCCCAA---ACGCUCAACGC---CACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACACAUACAUAUGC .((((.......))))...........(((.((((((((...(((((....---..)))))..((---(((.....(((......))).........)))))......)))))))).))) ( -31.54) >DroYak_CAF1 765 89 - 1 UGGCCAAAAAUCGGCCAUUACCA-------------UGUACAUAAGCUCAACGCUCGCUCGUCAG---CACCAAUCGAGCAUGUACA-------GUAGUGGCACGCACACAU-------- ...........((((((((((..-------------(((((((.(((.....))).(((((....---.......))))))))))))-------)))))))).)).......-------- ( -28.70) >consensus GGGCCAAAAAUCGGCCAUUACCA_CA______UGU_UGUACAUGAGCCCAA___ACGCUCGACAC___CACCAAUCGAGCAUGUACUCCACUAUAUAGUGGCACGCACACAUACAUAUG_ .((((.......)))).............................((((.......(((((..............)))))(((((......))))).).))).................. (-15.55 = -16.44 + 0.89)

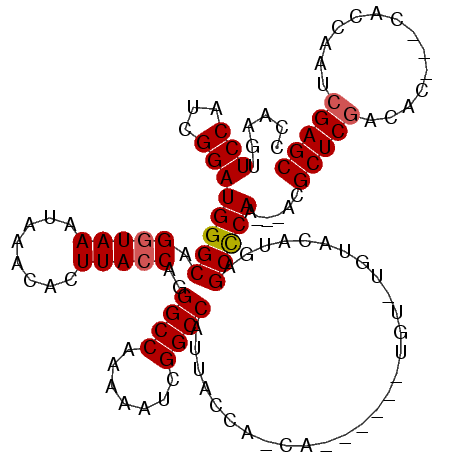
| Location | 1,665,564 – 1,665,675 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.41 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1665564 111 - 22224390 CCAAGUUCCAUCGGAUGGGCAGGUAAAUAAACACUUACCAGGGCCAAAAAUCGGCCAUUACCAUCA------UGUUUGUACAUGAGCCCAA---ACGCUCGACACCACCACCAAUCGAGC ............(..(((((.(((((........)))))..((((.......)))).......(((------(((....))))))))))).---.)((((((............)))))) ( -34.80) >DroSim_CAF1 763 114 - 1 CCAAGUUCCAUCGGAUGGGCAGGUAAAUAAACACUUACCAGGGCCAAAAAUCGGCCAUUACUACCAUGUACAUGUAUGUAAAUGAGCCCAA---ACGCUCAACGC---CACCAAUCGAGC ....((((.((.((...(((.(((((........)))))..((((.......)))).((((...(((....)))...)))).(((((....---..)))))..))---).)).)).)))) ( -30.00) >DroYak_CAF1 790 104 - 1 CAAAGUUCCAUCGGAUGGGCAGGUAAAUAAACACUUAACAUGGCCAAAAAUCGGCCAUUACCA-------------UGUACAUAAGCUCAACGCUCGCUCGUCAG---CACCAAUCGAGC ..........((((.(((((((((((.(((....)))...(((((.......)))))))))).-------------)))......(((..(((......))).))---).))).)))).. ( -24.20) >consensus CCAAGUUCCAUCGGAUGGGCAGGUAAAUAAACACUUACCAGGGCCAAAAAUCGGCCAUUACCA_CA______UGU_UGUACAUGAGCCCAA___ACGCUCGACAC___CACCAAUCGAGC ......(((...)))(((((.(((((........)))))..((((.......)))).............................)))))......(((((..............))))) (-22.96 = -23.41 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:52 2006