
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,330,325 – 15,330,439 |
| Length | 114 |
| Max. P | 0.966895 |

| Location | 15,330,325 – 15,330,439 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.36 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.93 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15330325 114 - 22224390 CUUGUACUUAGCUUCCUUGUUCCACGUUCAUAUCAAUCGGGCGUGCAAGU-----UAACAGUGGCGUUAACAUGCGUUAAGUACAGUACACCACACACACAACCCC-CGCUCCCCCUCCC .(((((((((((...........((((((.........))))))(((.((-----((((......)))))).))))))))))))))....................-............. ( -27.30) >DroPse_CAF1 18673 96 - 1 UCUAGCUUUGGUUUCCUUGUUCCAUAUCCAUAACAAUCAGGCGUGCAAAUGGAGUCGGCGUCAACGUUAACAUGCGUUAAGUACUGC--CCC--------AAC----CAC---------- ........(((((...(((((..........)))))...(((((((.((((.(((..(((....)))..)).).))))..)))).))--)..--------)))----)).---------- ( -19.50) >DroSec_CAF1 35065 97 - 1 CUUGUACUUAGCUUCCUUGUUCCACGUCCAUAUCAAUCGGGCGUGCAAGU-----UAACAGUGGCGUUAACAUGCGUUAAGUACAGUACCCC-------CAACCCU-UGC---------- .(((((((((((...........((((((.........))))))(((.((-----((((......)))))).))))))))))))))......-------.......-...---------- ( -30.10) >DroEre_CAF1 10863 95 - 1 CUUGUACUUAGCUUCCUUGUUCCACGUCCAUAUCAAUCGGGCGUGCAAGU-----UAACAGUGGCGUUAACAUGCGUUAAGUACAGUACCCC-------CUCCCCCU------------- .(((((((((((...........((((((.........))))))(((.((-----((((......)))))).))))))))))))))......-------........------------- ( -30.10) >DroYak_CAF1 11237 98 - 1 CUUGUACUUAGCUUCCUUGUUCCACGUCCAUAUCAAUCGGGCGUGCAAGU-----UAACAGUGGCGUUAACAUGCGUUAAGUACAGUACCCC-------CGCCCCUUUAC---------- .(((((((((((...........((((((.........))))))(((.((-----((((......)))))).))))))))))))))......-------...........---------- ( -30.10) >DroPer_CAF1 19506 96 - 1 UCUAGCUUUGGUUUCCUUGUUCCAUAUCCAUAACAAUCAGGCGUGCAAAUGGAGUCGGCGUCAACGUUAACAUGCGUUAAGUACUGC--CCC--------AAC----CAC---------- ........(((((...(((((..........)))))...(((((((.((((.(((..(((....)))..)).).))))..)))).))--)..--------)))----)).---------- ( -19.50) >consensus CUUGUACUUAGCUUCCUUGUUCCACGUCCAUAUCAAUCGGGCGUGCAAGU_____UAACAGUGGCGUUAACAUGCGUUAAGUACAGUACCCC_______CAACCC__CAC__________ .(((((((((.....((((...(((((((.........)))))))))))..............((((......))))))))))))).................................. (-13.85 = -14.93 + 1.09)

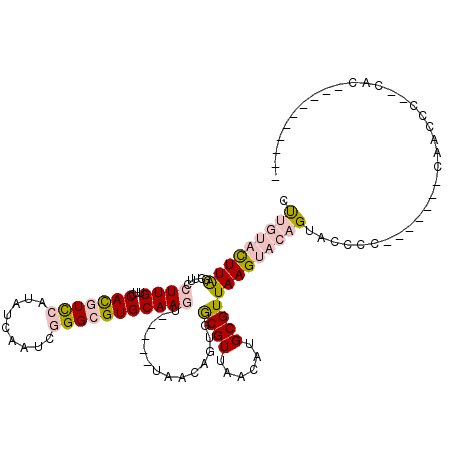
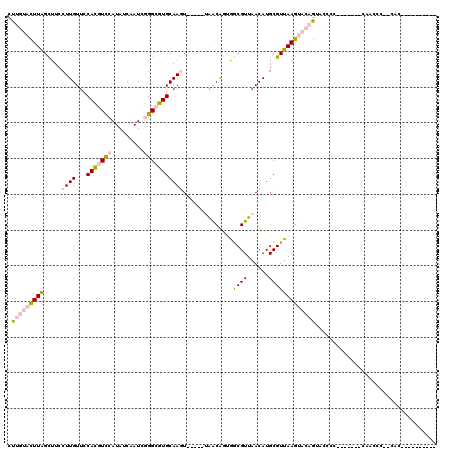
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:59 2006