
| Sequence ID | X_DroMel_CAF1 |
|---|---|
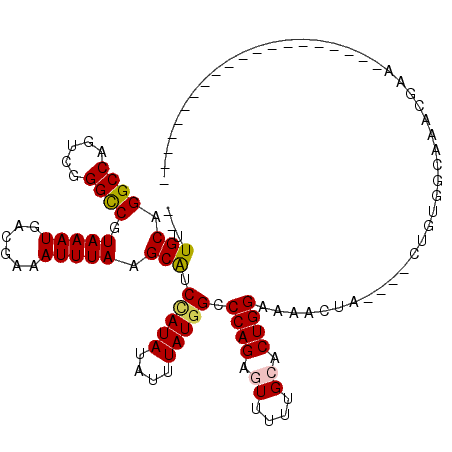
| Location | 15,327,791 – 15,327,885 |
| Length | 94 |
| Max. P | 0.587181 |

| Location | 15,327,791 – 15,327,885 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15327791 94 + 22224390 --UUGCAGGCCAGUCGGGCCGUAAAUGACGAAAUUUAAGCAUCUAUAUAUUUAUGGCCCAGAGUUUUUGCACUGGAAAACCA----CUGUGGCAAACGGA-------------------- --.......((....((((((((((((....................))))))))))))...(((((..((.(((....)))----.))..).)))))).-------------------- ( -26.45) >DroSim_CAF1 8348 94 + 1 --UUGCAGGCCAGUCGGGCCGUAAAUGACGAAAUUUAAGCAUCCAUAUAUUUAUGGCCCAGAGUUUUUGCACUGGAAAACUA----CUGUGGCAAACGAA-------------------- --(((...((((...((((((((((((....................))))))))))))(((((((((......))))))).----)).))))...))).-------------------- ( -27.05) >DroEre_CAF1 8400 94 + 1 --UUGCAGGCCAGUCGGGCCGUAAAUGACGAAAUUUAAGCAUCCAUAUAUUUAUGGCCCAGAGUUUUUGCACUGGAAAACUA----CUGUGGCAAACGAA-------------------- --(((...((((...((((((((((((....................))))))))))))(((((((((......))))))).----)).))))...))).-------------------- ( -27.05) >DroYak_CAF1 8108 94 + 1 --UUGCAGGCCAGUCGGGCCGUAAAUGACGAAAUUUAAGCAUCCAUAUAUUUAUGGCCCAGAGUUUUUGCACUGGAAAACUA----CUGUGGCAAUCGAA-------------------- --(((...((((...((((((((((((....................))))))))))))(((((((((......))))))).----)).))))...))).-------------------- ( -27.05) >DroAna_CAF1 29425 94 + 1 UUUUGCAGGCCAGUCGGGUCGUAAAUGACGAAAUUUAAGCGCCCAUAUAUUUAUGGCCCAGACUUUUUGCACUGGAAAAAUA----CUAAGA--AACAGG-------------------- ((((.(((((.((((((((((((((((....................)))))))))))).))))....)).))).))))...----......--......-------------------- ( -23.55) >DroPer_CAF1 14313 116 + 1 --UUGCAGGCCAGUCAGGCCAUAAAUGACGAAAUUUAAGCGGCCAUAUAUUUAUAGCCCAGACUUUUUGCACUGGAAAACUGAGUGCUAACA--GCCCGGGGUAGUGCAGGGGCAGGUGG --.(((.((((.....)))).(((((......))))).)))((((((....))).((((.......((((((((.....(((.(.((.....--))))))..)))))))))))).))).. ( -32.31) >consensus __UUGCAGGCCAGUCGGGCCGUAAAUGACGAAAUUUAAGCAUCCAUAUAUUUAUGGCCCAGAGUUUUUGCACUGGAAAACUA____CUGUGGCAAACGAA____________________ ...(((.((((.....)))).(((((......))))).))).(((((....))))).((((.((....)).))))............................................. (-18.54 = -18.57 + 0.03)


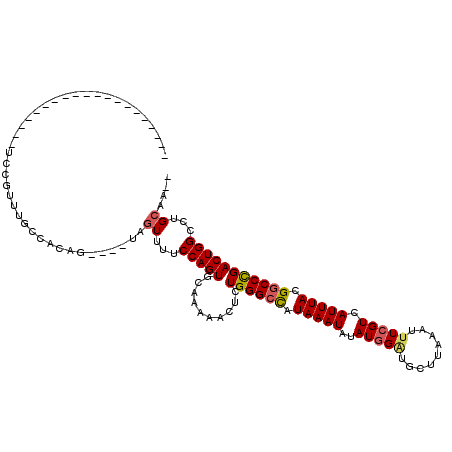
| Location | 15,327,791 – 15,327,885 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15327791 94 - 22224390 --------------------UCCGUUUGCCACAG----UGGUUUUCCAGUGCAAAAACUCUGGGCCAUAAAUAUAUAGAUGCUUAAAUUUCGUCAUUUACGGCCCGACUGGCCUGCAA-- --------------------...((..((((((.----(((....))).)).........((((((.(((((....((((......))))....))))).))))))..))))..))..-- ( -25.80) >DroSim_CAF1 8348 94 - 1 --------------------UUCGUUUGCCACAG----UAGUUUUCCAGUGCAAAAACUCUGGGCCAUAAAUAUAUGGAUGCUUAAAUUUCGUCAUUUACGGCCCGACUGGCCUGCAA-- --------------------...((..((((.((----.((((((........))))))))(((((.(((((..(((((.........))))).))))).)))))...))))..))..-- ( -25.10) >DroEre_CAF1 8400 94 - 1 --------------------UUCGUUUGCCACAG----UAGUUUUCCAGUGCAAAAACUCUGGGCCAUAAAUAUAUGGAUGCUUAAAUUUCGUCAUUUACGGCCCGACUGGCCUGCAA-- --------------------...((..((((.((----.((((((........))))))))(((((.(((((..(((((.........))))).))))).)))))...))))..))..-- ( -25.10) >DroYak_CAF1 8108 94 - 1 --------------------UUCGAUUGCCACAG----UAGUUUUCCAGUGCAAAAACUCUGGGCCAUAAAUAUAUGGAUGCUUAAAUUUCGUCAUUUACGGCCCGACUGGCCUGCAA-- --------------------...(...((((.((----.((((((........))))))))(((((.(((((..(((((.........))))).))))).)))))...))))...)..-- ( -22.90) >DroAna_CAF1 29425 94 - 1 --------------------CCUGUU--UCUUAG----UAUUUUUCCAGUGCAAAAAGUCUGGGCCAUAAAUAUAUGGGCGCUUAAAUUUCGUCAUUUACGACCCGACUGGCCUGCAAAA --------------------......--......----...((((.(((.((....((((.((((((((....)))))...........((((.....))))))))))).))))).)))) ( -20.40) >DroPer_CAF1 14313 116 - 1 CCACCUGCCCCUGCACUACCCCGGGC--UGUUAGCACUCAGUUUUCCAGUGCAAAAAGUCUGGGCUAUAAAUAUAUGGCCGCUUAAAUUUCGUCAUUUAUGGCCUGACUGGCCUGCAA-- .....(((....((.......(((((--(.((.(((((.........))))).)).))))))(((((((....)))))))))..................((((.....)))).))).-- ( -33.10) >consensus ____________________UCCGUUUGCCACAG____UAGUUUUCCAGUGCAAAAACUCUGGGCCAUAAAUAUAUGGAUGCUUAAAUUUCGUCAUUUACGGCCCGACUGGCCUGCAA__ ........................................((...(((((..........((((((.(((((..(((((.........))))).))))).)))))))))))...)).... (-16.40 = -16.68 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:58 2006