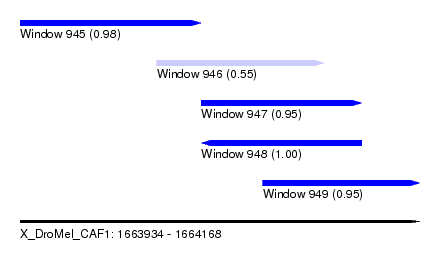
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,663,934 – 1,664,168 |
| Length | 234 |
| Max. P | 0.996632 |

| Location | 1,663,934 – 1,664,040 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -22.88 |
| Energy contribution | -25.12 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1663934 106 + 22224390 GGAAUUCCACAGCGAAACGGGCUGUAGUAAUGAAGAUAACAUUUCCUGUUGUCAGCAUUUGCAAAGUUGGACAAAGCCAGCUCGCAGCCC--CACACUCGCACCCUUG------------ ((....))...((((...(((((((.((((((..(((((((.....)))))))..)).))))..((((((......)))))).)))))))--.....)))).......------------ ( -36.60) >DroSim_CAF1 1897 118 + 1 GGAAUUCCAGAGCGAAACGGGCUGUAGUAAUGAAGAUAACAUUUCCUGUUGUCAGCAUUUGCAAAGUUGGACAAAGCCAGCUCGCAGCCC--CACAUCCGCACCCUUGCGUCCUCGCAUC ((....))...((((...(((((((.((((((..(((((((.....)))))))..)).))))..((((((......)))))).)))))))--......((((....))))...))))... ( -40.90) >DroEre_CAF1 9619 80 + 1 GGAAUUCCAAAGCGAAACGGGCUGUAGUAAUGAAGAUAACAUUUCCUGUUGUCAGCAUUUGCAAAGUUGGACAAAGCCAG---------------------------------------- ((...(((((.(((((....(((.((....))..(((((((.....)))))))))).)))))....))))).....))..---------------------------------------- ( -18.50) >DroYak_CAF1 2119 116 + 1 GGAAUUCCAGAGCGAAACGGGCUGUAGUAAUGAAGAUAACAUUUCCUGUUGUCAGCAUUUGCAAAGUUGGACAAAGCCAGCUCGCAGCCCCCCACAUCCACAUCCA--CAUC--CGCAUC (((.(((......)))..(((((((.((((((..(((((((.....)))))))..)).))))..((((((......)))))).)))))))......))).......--....--...... ( -32.90) >consensus GGAAUUCCAGAGCGAAACGGGCUGUAGUAAUGAAGAUAACAUUUCCUGUUGUCAGCAUUUGCAAAGUUGGACAAAGCCAGCUCGCAGCCC__CACAUCCGCACCCU______________ ((....))..........(((((((.((((((..(((((((.....)))))))..)).))))..((((((......)))))).))))))).............................. (-22.88 = -25.12 + 2.25)

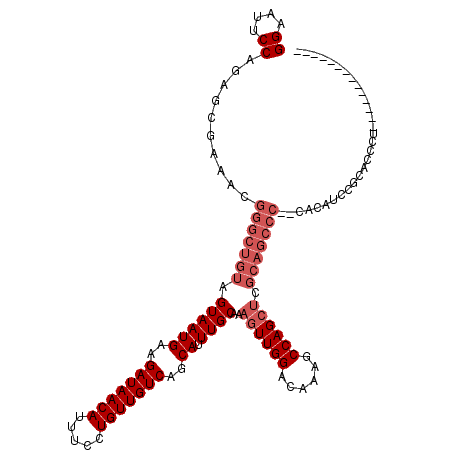
| Location | 1,664,014 – 1,664,112 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.73 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1664014 98 + 22224390 CUCGCAGCCC--CACACUCGCACCCUUG----------------CAUCCUGGUAUCCUGGCACUCCACGCGCUUCAAACUUUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGGA .........(--(......(((....))----------------).((((..(((..((((((((((...((((((.......))))))))-).)))))))..)))..).))).)). ( -29.00) >DroSim_CAF1 1977 112 + 1 CUCGCAGCCC--CACAUCCGCACCCUUGCGUCCUCGCAUCCUCGCAUCCUCGCAUCCUGGCACUCC--GCGCUUCAAACUUUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGGA ..........--....((((((....)))((((((((......((......))....(((((((((--(.((((((.......))))))))-).)))))))..))))...))))))) ( -38.80) >DroYak_CAF1 2199 111 + 1 CUCGCAGCCCCCCACAUCCACAUCCA--CAUC--CGCAUCCUCGCAUCCUCGCACCCUCGCACUUC--UCGCUUCAAAGUUUAUGAAGCUGGGAAGUGGCAAAAUGAAACGGACGGA ..........................--..((--((..(((..((......))......(((((((--((((((((.......)))))).))))))).))..........))))))) ( -27.50) >consensus CUCGCAGCCC__CACAUCCGCACCCUUGC_UC__CGCAUCCUCGCAUCCUCGCAUCCUGGCACUCC__GCGCUUCAAACUUUAUGAAGCUG_GAAGUGCCAAAGUGAAACGGACGGA .......................((..........((......)).(((...(((..(((((((.(....((((((.......))))))...).)))))))..)))....))).)). (-17.54 = -18.73 + 1.20)

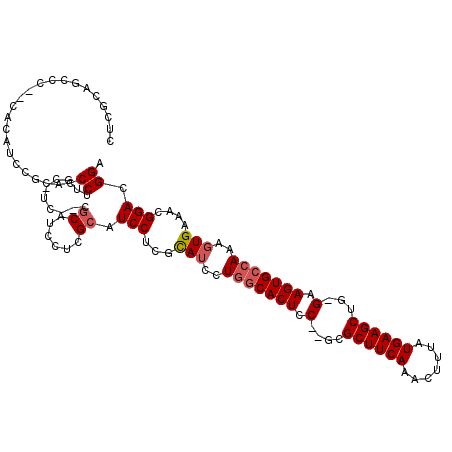
| Location | 1,664,040 – 1,664,134 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -21.11 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1664040 94 + 22224390 ----CAUCCUGGUAUCCUGGCACUCCACGCGCUUCAAACUUUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGGACUGUAACAGUUUUUCGAUUGCU------------------ ----.............((((((((((...((((((.......))))))))-).))))))).((..(..((((.((((((...)))))))))).)..))------------------ ( -28.30) >DroSim_CAF1 2015 96 + 1 CUCGCAUCCUCGCAUCCUGGCACUCC--GCGCUUCAAACUUUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGGACUGUAACAGUUUUUCGAUUGUU------------------ ...((((((((.(((..(((((((((--(.((((((.......))))))))-).)))))))..))).....)).))).)))(((((((....)))))))------------------ ( -32.00) >DroYak_CAF1 2235 115 + 1 CUCGCAUCCUCGCACCCUCGCACUUC--UCGCUUCAAAGUUUAUGAAGCUGGGAAGUGGCAAAAUGAAACGGACGGACUGUUACAGUUUUUCGAUUGCUGAAAAAGGAGCGAAAGUU .((((.((((..........((((((--((((((((.......)))))).))))))))((((...(((((.(((.....)))...)))))....))))......))))))))..... ( -36.00) >consensus CUCGCAUCCUCGCAUCCUGGCACUCC__GCGCUUCAAACUUUAUGAAGCUG_GAAGUGCCAAAGUGAAACGGACGGACUGUAACAGUUUUUCGAUUGCU__________________ ...........(((...(((((((.(....((((((.......))))))...).)))))))........((((.((((((...))))))))))..)))................... (-21.11 = -21.33 + 0.23)

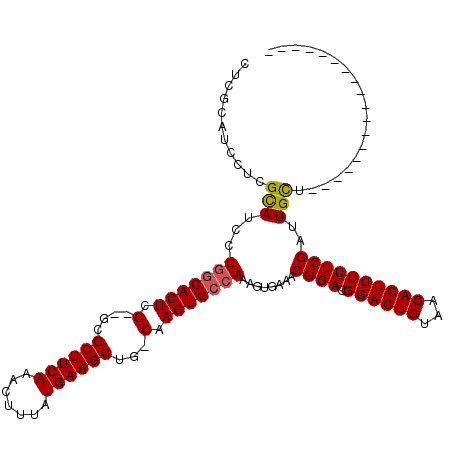
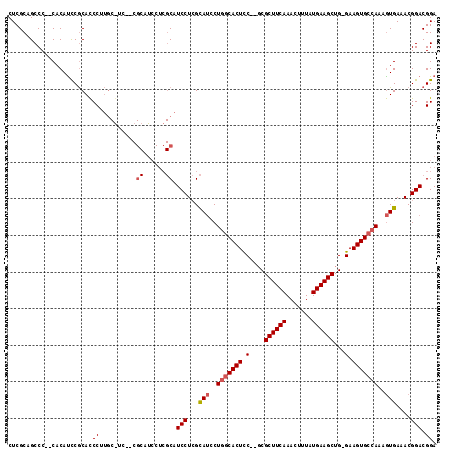
| Location | 1,664,040 – 1,664,134 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -28.35 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1664040 94 - 22224390 ------------------AGCAAUCGAAAAACUGUUACAGUCCGUCCGUUUCACUUUGGCACUUC-CAGCUUCAUAAAGUUUGAAGCGCGUGGAGUGCCAGGAUACCAGGAUG---- ------------------............((((...)))).(((((.......(((((((((((-(.((((((.......))))))..).)))))))))))......)))))---- ( -30.62) >DroSim_CAF1 2015 96 - 1 ------------------AACAAUCGAAAAACUGUUACAGUCCGUCCGUUUCACUUUGGCACUUC-CAGCUUCAUAAAGUUUGAAGCGC--GGAGUGCCAGGAUGCGAGGAUGCGAG ------------------.....(((....((((...)))).(((((.((.((..((((((((((-..((((((.......))))))..--))))))))))..)).)))))))))). ( -34.60) >DroYak_CAF1 2235 115 - 1 AACUUUCGCUCCUUUUUCAGCAAUCGAAAAACUGUAACAGUCCGUCCGUUUCAUUUUGCCACUUCCCAGCUUCAUAAACUUUGAAGCGA--GAAGUGCGAGGGUGCGAGGAUGCGAG .....(((((((((.....(((..(((...((((...))))...)).........((((.(((((...((((((.......))))))..--))))))))))..)))))))).)))). ( -33.80) >consensus __________________AGCAAUCGAAAAACUGUUACAGUCCGUCCGUUUCACUUUGGCACUUC_CAGCUUCAUAAAGUUUGAAGCGC__GGAGUGCCAGGAUGCGAGGAUGCGAG ..............................((((...)))).(((((....((..((((((((((...((((((.......))))))....))))))))))..))...))))).... (-28.35 = -29.13 + 0.78)

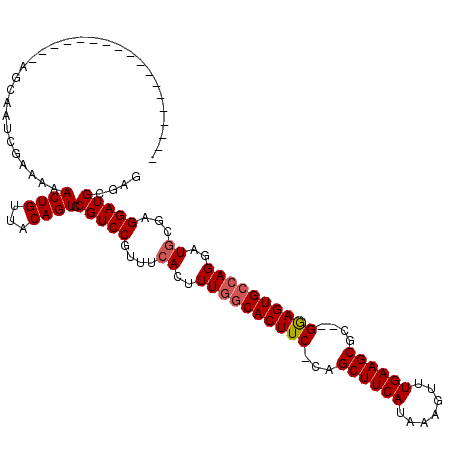
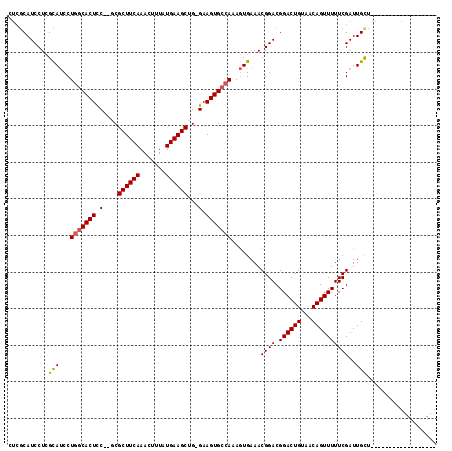
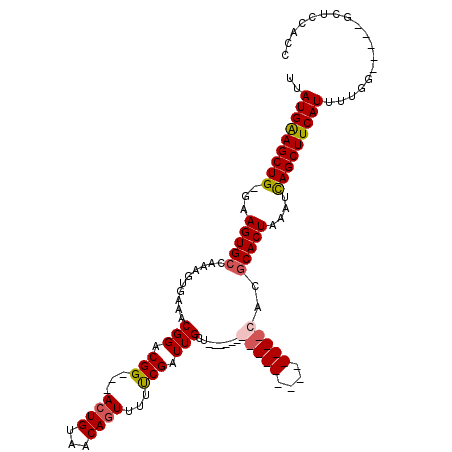
| Location | 1,664,076 – 1,664,168 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.15 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1664076 92 + 22224390 UUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGG---ACUGUAACAGUUUUUCGAUUGCU-------------------CUUGCACUAAAUCAGCUUCAUUUUGG-----GCUCCACC ..(((((((((-(.(((((...((..(..((((.((---((((...)))))))))).)..))-------------------...)))))...))))))))))...((-----......)) ( -28.10) >DroSim_CAF1 2053 92 + 1 UUAUGAAGCUG-GAAGUGCCAAAGUGAAACGGACGG---ACUGUAACAGUUUUUCGAUUGUU-------------------CUUGCACUAAAUCAGCUUCAUUUUGG-----GCUCCACC ..(((((((((-(.(((((..(((...((((((.((---((((...)))))))))).))...-------------------))))))))...))))))))))...((-----......)) ( -26.10) >DroEre_CAF1 9725 113 + 1 UUAUGGAGCUG-GAAGUGCCAAAGUGAAACGGACGGACGACUGUUACACUUUUCCGAUUGCUCAA-AAGCAGCGAAAGUUCCACGCACUAAAUCAGCUUCAUUUUUG-----GCUCCGCU ....(((((..-((((((..((((((.(((((.(....).))))).))))))...(((((((...-.)))(((....)))..........)))).....))))))..-----)))))... ( -34.50) >DroYak_CAF1 2273 117 + 1 UUAUGAAGCUGGGAAGUGGCAAAAUGAAACGGACGG---ACUGUUACAGUUUUUCGAUUGCUGAAAAAGGAGCGAAAGUUCCACGCACUAAAUUAGCUUCAUUUUUGGGGGAGCUCUACU ..((((((((((..((((((((...(((((.(((..---...)))...)))))....)))).......(((((....)))))...))))...))))))))))....(((....))).... ( -35.80) >consensus UUAUGAAGCUG_GAAGUGCCAAAGUGAAACGGACGG___ACUGUAACAGUUUUUCGAUUGCU___________________CACGCACUAAAUCAGCUUCAUUUUGG_____GCUCCACC ..(((((((((...(((((..........(((.(((...((((...))))...))).)))........(((((....)))))..)))))....))))))))).................. (-19.96 = -21.15 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:48 2006