| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,305,330 – 15,305,433 |
| Length | 103 |
| Max. P | 0.837436 |

| Location | 15,305,330 – 15,305,433 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 72.01 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -18.68 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15305330 103 - 22224390 AGAGGAACAGGCGAAACUGGAGGCAGCCAAG-AAUACCUCCGAAACGGAGGUGGACUGGGCAGCGCUGCUGUCCGAGGAUUUUCUGGAUCCGCCAGAGGAUGAC .........((((...(..((((...((.((-..((((((((...))))))))..))(((((((...)))))))..))..))))..)...)))).......... ( -39.00) >DroSec_CAF1 389 103 - 1 ACAGGAACAGGCGAAACUGGAGGCAACCAGG-AAUACCUCCGAAACGGAUGAGGAUUGGCCAGCGCUGCUGGCCGAGGAUUUCCUGGAUCCGCCUGAGGAUGAC .((....((((((....((....)).(((((-((..((((((...))))......(((((((((...)))))))))))..)))))))...))))))....)).. ( -41.80) >DroSim_CAF1 749 103 - 1 ACAGGAACAGGCGAAACUGGAGGCAACCAGG-AAUACCUCCGAAACGGAGGUGGACUGGCCAGCGCUGCUGGCCGAGGAUUUCCUGGAUCCGCCUGAGGAUGAC .((....((((((....((....)).(((((-((((((((((...))))))))..(((((((((...))))))).))...)))))))...))))))....)).. ( -47.30) >DroEre_CAF1 672 93 - 1 -------GAGGUAGAACUGGAGGCCGCCAUG-CAUCCAGACGA---GGAGGUGGACUGGGCAGCGUUGCUGUCCGAGGAUUUCCUGGAUCCACCAGAGUAUGAC -------..(((..........)))..((((-(.(((......---)))(((((((((((((((...))))))).(((....))))).))))))...))))).. ( -34.50) >DroYak_CAF1 725 96 - 1 -------CAGGUGAAACUGGAGGUAGCCGUC-AAUUCCGCCGAAACGGAGGUGGACUGGGCAGCCCUGCUAGCCGAGGAUUUCCUGGAUCCGCCAGACGAUGAC -------..((((...(((((((..((.(((-...((((((........))))))...))).))))).))))((.(((....)))))...)))).......... ( -32.90) >DroAna_CAF1 702 97 - 1 -------CAGUGGCGCCUGCACACCGUCAAGCAAUGCUGCCGAGGGAGAAAUCGACUGGGUGGCGUUGCUAAGCGAGGAUUUUGUUCCACCGCUUUCCCCGGGC -------.......(((((..........((((((((..((.((.((....))..)).))..))))))))(((((.(((......)))..)))))....))))) ( -39.50) >consensus _______CAGGCGAAACUGGAGGCAGCCAAG_AAUACCGCCGAAACGGAGGUGGACUGGGCAGCGCUGCUGGCCGAGGAUUUCCUGGAUCCGCCAGAGGAUGAC .........((((.....(((((...((........(((((........)))))..((((((((...)))))))).)).)))))......)))).......... (-18.68 = -20.13 + 1.45)

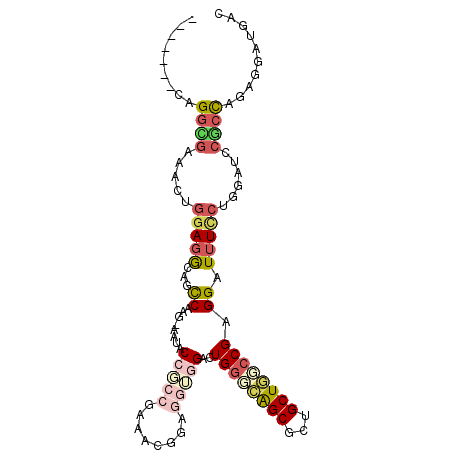

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:49 2006