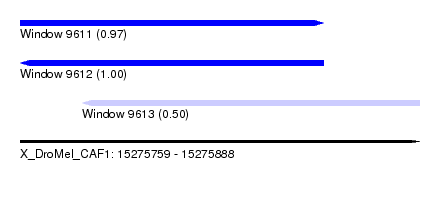
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,275,759 – 15,275,888 |
| Length | 129 |
| Max. P | 0.996274 |

| Location | 15,275,759 – 15,275,857 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -29.66 |
| Energy contribution | -30.33 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15275759 98 + 22224390 GAGCCGAAAGGACACUACUACUGCUCCUCAGGCGGAACAGCCCGCUGGGAAAUGGAGCAGUAGUGUCCGGGUUUUUGUUUUUUUGUGGGGCGGGGAUC .........(((((((...((((((((((.(((((......)))))..))...))))))))))))))).((((((((((((.....)))))))))))) ( -40.70) >DroSec_CAF1 99692 94 + 1 GAGCCGAAAGGACACUACUACUGCUCCUCAGGCGGAACAGCCCGCUGGGAAAUGGAGCAGUAGUGUCCGGGUUUUUUUUUUUU----GGGAGGGAAUC (((((....(((((((...((((((((((.(((((......)))))..))...))))))))))))))).)))))(((((((..----..))))))).. ( -38.00) >DroYak_CAF1 87958 87 + 1 GAGCCGAAAGGACACUACAACUGCUCCUUAGGCGAAACAGCCCGUUGGGAAAUGGAGCAAUAGUAUUCGGGUAUUUU-----U----GGCA--AAUUC ..((((((((.((..(((...((((((...(((......))).(((....)))))))))...))).....)).))))-----)----))).--..... ( -24.70) >consensus GAGCCGAAAGGACACUACUACUGCUCCUCAGGCGGAACAGCCCGCUGGGAAAUGGAGCAGUAGUGUCCGGGUUUUUUUUUUUU____GGGAGGGAAUC (((((....(((((((((...((((((((.(((((......)))))..))...))))))))))))))).)))))........................ (-29.66 = -30.33 + 0.67)



| Location | 15,275,759 – 15,275,857 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -27.67 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15275759 98 - 22224390 GAUCCCCGCCCCACAAAAAAACAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAGUAGUAGUGUCCUUUCGGCUC .......(((....................(((((((((((((....(((((((((((....)))).))).)))).)))))))))))))....))).. ( -33.95) >DroSec_CAF1 99692 94 - 1 GAUUCCCUCCC----AAAAAAAAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAGUAGUAGUGUCCUUUCGGCUC ...........----...............(((((((((((((....(((((((((((....)))).))).)))).)))))))))))))......... ( -33.70) >DroYak_CAF1 87958 87 - 1 GAAUU--UGCC----A-----AAAAUACCCGAAUACUAUUGCUCCAUUUCCCAACGGGCUGUUUCGCCUAAGGAGCAGUUGUAGUGUCCUUUCGGCUC .....--.(((----.-----(((..((.....(((.((((((((.((.......((((......)))))))))))))).)))..))..))).))).. ( -21.31) >consensus GAUUCCCUCCC____AAAAAAAAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAGUAGUAGUGUCCUUUCGGCUC ..............................(((((((((((((....((((...(((((......))))).)))).)))))))))))))......... (-27.67 = -28.23 + 0.56)


| Location | 15,275,779 – 15,275,888 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -18.60 |
| Energy contribution | -17.83 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15275779 109 - 22224390 UCAAUUUGCUUCUAUAACACGCCUAAUCUGUGAUCCCCGCCCCACAAAAAAACAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAG ......(((((((......(((......((((..........))))...............(((((....)).)))........)))(((......)))..))))))). ( -21.50) >DroSec_CAF1 99712 105 - 1 UCAAUUUGCUUCUAUAACACGCCUAAUCUGUGAUUCCCUCCC----AAAAAAAAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAG ......(((((((....((((.......))))..........----..............((((.((..(((((.........)))))...))))))....))))))). ( -20.90) >DroYak_CAF1 87978 98 - 1 UCAAUUUGCUUCUAUAAUACGCCUAAUGCGUGAAUU--UGCC----A-----AAAAUACCCGAAUACUAUUGCUCCAUUUCCCAACGGGCUGUUUCGCCUAAGGAGCAG ......(((((((....(((((.....)))))....--.((.----.-----.(((((((((.......................)))).))))).))...))))))). ( -19.80) >consensus UCAAUUUGCUUCUAUAACACGCCUAAUCUGUGAUUCCCUCCC____AAAAAAAAAAAACCCGGACACUACUGCUCCAUUUCCCAGCGGGCUGUUCCGCCUGAGGAGCAG ......(((((((....((((.......))))............................((((.((..(((((.........)))))...))))))....))))))). (-18.60 = -17.83 + -0.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:41 2006