| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,662,394 – 1,662,507 |
| Length | 113 |
| Max. P | 0.895693 |

| Location | 1,662,394 – 1,662,507 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -24.98 |
| Energy contribution | -26.60 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1662394 113 + 22224390 UUCGAGUGGAUGUGUCGCGGCAACUUAAGUGUAUCAUU------UGCUUGGCCAGAAUCA-GGCGGCAAUUGAUGGGGCUUCAUUGAAGCCGCGAAAGUGUUGCGGAAUUAAGUUACUCG ..(((((((..((..(((((((.(((..((.(((((.(------((((..(((.......-)))))))).))))).((((((...))))))))..))))))))))..))....))))))) ( -42.00) >DroSim_CAF1 405 117 + 1 UUGGAGUGGAUGUGUCGCGGCAACUUAAGUGUAUCAUUGCUUGGUGCUUGGCCAGCACCA-GGCGGCAAUUGAUGGGGCUUCAUUGAAGCCGCGAAAGUGUUGCGAAAUUAAGUUACU-- ....(((((..((.((((((((.(((..((.(((((((((((((((((.....)))))))-)))))....))))).((((((...))))))))..))))))))))).))....)))))-- ( -44.60) >DroEre_CAF1 8120 101 + 1 UUCGAGUGGCUGUGUCG------------UGUAUCUUU------UGCUCGGCCAGCAGCAAAGCGGCCAUUGAUGGGGUUUCACUGAAGCCGCGAAAGUGUUGCGAA-CCAACUUACUCG ..(((((((.((..(((------------((...((((------(((..((((.((......))))))........((((((...)))))))))))))...))))).-.)).).)))))) ( -37.20) >DroYak_CAF1 567 113 + 1 UUCGAGUGGAUGUGUCGCGGCAACUUAAGUGUAUCAUU------UGCUUGGCCAGCAGCA-AGCGGCAAUUGAUGGGGUUUUAUUGAAGCCGCGAAAGUGGUGCGAAAUGAAGUUACUCG ..(((((((.....((((..(((...((.(.(((((.(------((((..((.....)).-...))))).))))).).))...)))..(((((....))))))))).......))))))) ( -36.30) >consensus UUCGAGUGGAUGUGUCGCGGCAACUUAAGUGUAUCAUU______UGCUUGGCCAGCAGCA_AGCGGCAAUUGAUGGGGCUUCAUUGAAGCCGCGAAAGUGUUGCGAAAUUAAGUUACUCG ..(((((((.....((((((((............................(((.((......)))))......((.((((((...)))))).))....)))))))).......))))))) (-24.98 = -26.60 + 1.62)

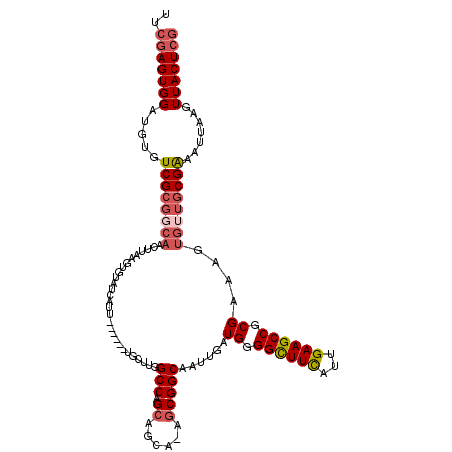
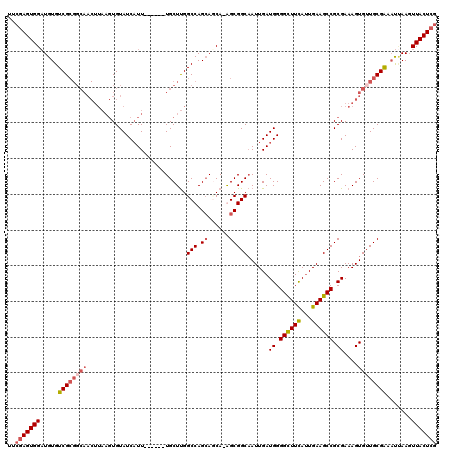
| Location | 1,662,394 – 1,662,507 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1662394 113 - 22224390 CGAGUAACUUAAUUCCGCAACACUUUCGCGGCUUCAAUGAAGCCCCAUCAAUUGCCGCC-UGAUUCUGGCCAAGCA------AAUGAUACACUUAAGUUGCCGCGACACAUCCACUCGAA (((((........((((((((......(.((((((...)))))).)((((.((((.(((-.......)))...)))------).))))........)))).)).)).......))))).. ( -30.56) >DroSim_CAF1 405 117 - 1 --AGUAACUUAAUUUCGCAACACUUUCGCGGCUUCAAUGAAGCCCCAUCAAUUGCCGCC-UGGUGCUGGCCAAGCACCAAGCAAUGAUACACUUAAGUUGCCGCGACACAUCCACUCCAA --.(((((((((....((((.......(.((((((...)))))))......)))).((.-(((((((.....))))))).))..........)))))))))................... ( -33.12) >DroEre_CAF1 8120 101 - 1 CGAGUAAGUUGG-UUCGCAACACUUUCGCGGCUUCAGUGAAACCCCAUCAAUGGCCGCUUUGCUGCUGGCCGAGCA------AAAGAUACA------------CGACACAGCCACUCGAA (((((..((((.-.(((.....((((..(((((.(((..((...(((....)))....))..)))..)))))....------)))).....------------)))..)))).))))).. ( -29.90) >DroYak_CAF1 567 113 - 1 CGAGUAACUUCAUUUCGCACCACUUUCGCGGCUUCAAUAAAACCCCAUCAAUUGCCGCU-UGCUGCUGGCCAAGCA------AAUGAUACACUUAAGUUGCCGCGACACAUCCACUCGAA (((((.........((((.(.((((..(((((.....................)))))(-((((........))))------)...........)))).)..)))).......))))).. ( -23.99) >consensus CGAGUAACUUAAUUUCGCAACACUUUCGCGGCUUCAAUGAAACCCCAUCAAUUGCCGCC_UGCUGCUGGCCAAGCA______AAUGAUACACUUAAGUUGCCGCGACACAUCCACUCGAA (((((.........((((.........(((((....(((......))).....))))).....((((.....))))..........................)))).......))))).. (-16.20 = -17.52 + 1.31)

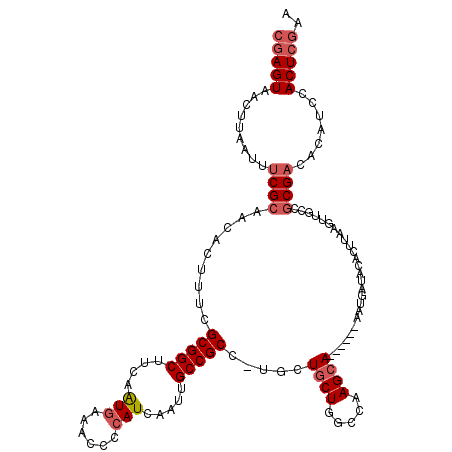
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:44 2006