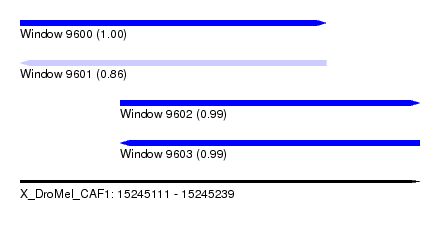
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,245,111 – 15,245,239 |
| Length | 128 |
| Max. P | 0.998091 |

| Location | 15,245,111 – 15,245,209 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15245111 98 + 22224390 UGUACAUGGGUAUAUAUGUAUGUAUAUAGAUAUCUCUAUGCCGACACCCAAGUUGAGGUGCCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU ......(((((......((..(((((.((....)).)))))..)))))))(((((((((((((((...........)))).)))))))))))...... ( -26.40) >DroSec_CAF1 65381 96 + 1 UGUGCAUGGGUAUAUAUGUAUGUACAUAGAUC--UCUAUGCCGACACCCAAGUUGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU .(.((((((..((.(((((....))))).)).--.)))))))(((((((.....).)))))).........(((((((((.....))))))))).... ( -30.40) >DroSim_CAF1 74218 96 + 1 CGUGCAUGGGUAUAUAUGUAUGUAUAUAGAUA--UCUAUGCCGACACCCAAGUUGAGGUGCCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU ((.((((((((((.(((((....))))).)))--))))))))).......(((((((((((((((...........)))).)))))))))))...... ( -32.80) >DroEre_CAF1 66123 74 + 1 ----UAUACAUAG------------------C--UCUAUGCCGACACCCAAGCGGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU ----....((((.------------------.--..))))..((((((........)))))).........(((((((((.....))))))))).... ( -19.30) >DroYak_CAF1 65830 78 + 1 UGUGUAUAGGUAU------------------A--UCUAUGCUGACACCCAAGUCGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU ...(((((((...------------------.--))))))).((((((........)))))).........(((((((((.....))))))))).... ( -23.40) >consensus UGUGCAUGGGUAUAUAUGUAUGUA_AUAGAUA__UCUAUGCCGACACCCAAGUUGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAU ...(((((((....(((((....)))))......))))))).(((((((.....).)))))).........(((((((((.....))))))))).... (-18.96 = -19.74 + 0.78)


| Location | 15,245,111 – 15,245,209 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15245111 98 - 22224390 AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGGCACCUCAACUUGGGUGUCGGCAUAGAGAUAUCUAUAUACAUACAUAUAUACCCAUGUACA ....((.(((((....(((......))).)))))..(((((((......))))))).))(((((....))))).....(((((........))))).. ( -22.60) >DroSec_CAF1 65381 96 - 1 AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCAACUUGGGUGUCGGCAUAGA--GAUCUAUGUACAUACAUAUAUACCCAUGCACA ....((((((((....(((......))).)))))..(((((((......))))))).((((((.--...)))))).................)))... ( -25.40) >DroSim_CAF1 74218 96 - 1 AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGGCACCUCAACUUGGGUGUCGGCAUAGA--UAUCUAUAUACAUACAUAUAUACCCAUGCACG ....(((....((((((((......)))....(((.(((((((......)))))))..)))...--)))))(((((......))))).....)))... ( -21.50) >DroEre_CAF1 66123 74 - 1 AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCCGCUUGGGUGUCGGCAUAGA--G------------------CUAUGUAUA---- ..((((......(((.(((......)))))).(((.(((((((......)))))))..)))...--)------------------)))......---- ( -20.60) >DroYak_CAF1 65830 78 - 1 AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCGACUUGGGUGUCAGCAUAGA--U------------------AUACCUAUACACA ....((.(((((....(((......))).)))))..(((((((......))))))).))((((.--.------------------....))))..... ( -19.90) >consensus AUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCAACUUGGGUGUCGGCAUAGA__UAUCUAU_UACAUACAUAUAUACCCAUGCACA ....((.(((((....(((......))).)))))..(((((((......))))))).))....................................... (-18.64 = -18.40 + -0.24)


| Location | 15,245,143 – 15,245,239 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -20.24 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15245143 96 + 22224390 UCUCUAUGCCGACACCCAAGUUGAGGUGCCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAACCCGAAACAGGGCCAAA ......(((.........(((((((((((((((...........)))).))))))))))).........))).......(((......)))..... ( -23.97) >DroSec_CAF1 65413 94 + 1 --UCUAUGCCGACACCCAAGUUGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAACACGAAACAGGGCCAAA --.....((((((((((.....).)))))).........(((((((((.....)))))))))...........................))).... ( -23.20) >DroSim_CAF1 74250 94 + 1 --UCUAUGCCGACACCCAAGUUGAGGUGCCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAACCCGAAACAGGGCCAAA --....(((.........(((((((((((((((...........)))).))))))))))).........))).......(((......)))..... ( -23.97) >DroEre_CAF1 66133 94 + 1 --UCUAUGCCGACACCCAAGCGGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAACCCGAAACAGGGUCAAA --....(((.((((((........)))))).........(((((((((.....))))))))).......)))......((((......)))).... ( -26.10) >DroYak_CAF1 65844 94 + 1 --UCUAUGCUGACACCCAAGUCGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAAGCCGAAACAGGGCCAAA --....(((.((((((........)))))).........(((((((((.....))))))))).......))).......(((.......))).... ( -24.10) >consensus __UCUAUGCCGACACCCAAGUUGAGGUGUCUCAUCAAUCGCAAUUGAGCGCAUCUUAAUUGCUAAUUAUGCAAAUUCAACCCGAAACAGGGCCAAA .......((((((((((.....).)))))).........(((((((((.....)))))))))...........................))).... (-20.24 = -20.32 + 0.08)



| Location | 15,245,143 – 15,245,239 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -26.96 |
| Energy contribution | -26.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15245143 96 - 22224390 UUUGGCCCUGUUUCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGGCACCUCAACUUGGGUGUCGGCAUAGAGA ((..((((......))))..))..(((.(((((.((((((.((.....)).)))))))))))....(((((((......))))))).)))...... ( -29.10) >DroSec_CAF1 65413 94 - 1 UUUGGCCCUGUUUCGUGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCAACUUGGGUGUCGGCAUAGA-- ....(((.....(((((((((...(((((....(....)....))))).)))).))))).......(((((((......)))))))))).....-- ( -25.00) >DroSim_CAF1 74250 94 - 1 UUUGGCCCUGUUUCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGGCACCUCAACUUGGGUGUCGGCAUAGA-- ((..((((......))))..))..(((.(((((.((((((.((.....)).)))))))))))....(((((((......))))))).)))....-- ( -29.10) >DroEre_CAF1 66133 94 - 1 UUUGACCCUGUUUCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCCGCUUGGGUGUCGGCAUAGA-- ((..((((......))))..))..(((.(((((.((((((.((.....)).)))))))))))....(((((((......))))))).)))....-- ( -29.40) >DroYak_CAF1 65844 94 - 1 UUUGGCCCUGUUUCGGCUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCGACUUGGGUGUCAGCAUAGA-- ((.((((.......)))).))...(((.(((((.((((((.((.....)).)))))))))))....(((((((......))))))).)))....-- ( -24.60) >consensus UUUGGCCCUGUUUCGGGUUGAAUUUGCAUAAUUAGCAAUUAAGAUGCGCUCAAUUGCGAUUGAUGAGACACCUCAACUUGGGUGUCGGCAUAGA__ ((..((((......))))..))..(((.(((((.((((((.((.....)).)))))))))))....(((((((......))))))).)))...... (-26.96 = -26.96 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:32 2006