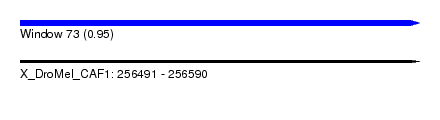
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 256,491 – 256,590 |
| Length | 99 |
| Max. P | 0.949910 |

| Location | 256,491 – 256,590 |
|---|---|
| Length | 99 |
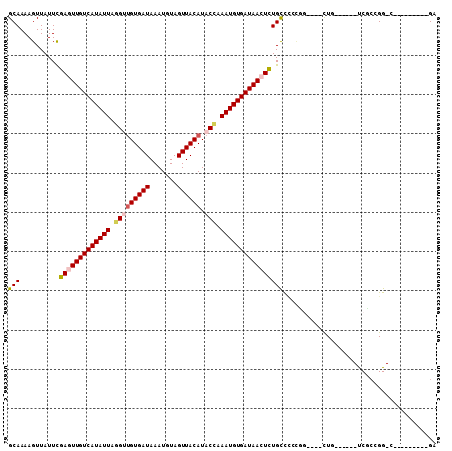
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
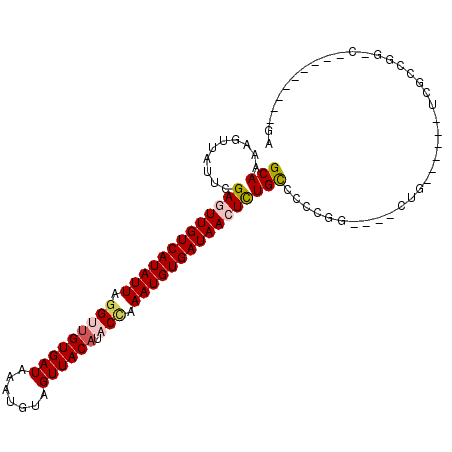
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -18.61 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 256491 99 + 22224390 GCAAAAGUUAUUUGAGUUGUCAUAUUAGGCCGUGAUAAAUGUAGUUACAUACCAAAUGUGAUAACUCUGCCCCUGG----CUU-UGGAUUCCCCGGAC-----CCUGGA .............(((((((((((((.((..(((((.......)))))...)).)))))))))))))....((.((----.((-(((.....))))).-----)).)). ( -29.40) >DroVir_CAF1 5244 86 + 1 GCAAAAGUUAUUCGACUUGUCAUAUUAGGUUGUGAUAAAUGUAGUUACAUACCCAAUGUGAUAACUUUGCCCG-------CUG------UCGCCGG-C---------GC (((((((((....))))(((((((((.(((((((((.......)))))).))).)))))))))..))))).((-------(((------....)))-)---------). ( -25.40) >DroGri_CAF1 17112 89 + 1 GCAAAAGUUAUUCGAGUUGUCAUAUUAGGUUGUGAUAAAUGUAGUUACAUACCCAAUGUGAUAACUUUGCUCGCCG----CUG------UCGCCGG-C---------GC (((..........(((((((((((((.(((((((((.......)))))).))).)))))))))))))))).(((((----(..------..).)))-)---------). ( -30.10) >DroEre_CAF1 3570 98 + 1 GCAAAAGUUAUUUGAGUUGUCAUAUUAGGCUGUGAUAAAUGUAGUUACAUACCAAAUGUGAUAACUCUGCC-CUGG----CUU-UGGAUUCCCCGGAC-----UCUGGA ...(((((((...(((((((((((((.((.((((((.......))))))..)).)))))))))))))....-.)))----)))-)(((.((....)).-----)))... ( -28.40) >DroAna_CAF1 4420 103 + 1 GCAAAAGUUAUUUGAGUUGUCAUAUUAAGUUGUGAUAAAUGUAGUUACAUACCAAAUGUGAUAACUCUGUCUCCGA----UUUCUCGAUUCU--GGACUUGGACCUGGA .............(((((((((((((..((((((((.......)))))).))..)))))))))))))....(((((----.(((........--))).)))))...... ( -26.10) >DroMoj_CAF1 5281 93 + 1 GCAAAAGUUAUUCGACUUGUCAUAUUAGGUUGUGAUAAAUGUAGUUACAUACUCAAUGUGAUAACUUUGCUCGCCGAUUGCUG------CCGCCGG-C---------GC (((((((((....))))(((((((((..((((((((.......)))))).))..)))))))))..))))).(((((...((..------..)))))-)---------). ( -26.90) >consensus GCAAAAGUUAUUCGAGUUGUCAUAUUAGGUUGUGAUAAAUGUAGUUACAUACCAAAUGUGAUAACUCUGCCCCCGG____CUG______UCGCCGG_C_________GA (((..........(((((((((((((.(((((((((.......)))))).))).))))))))))))))))....................................... (-18.61 = -19.12 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:37 2006