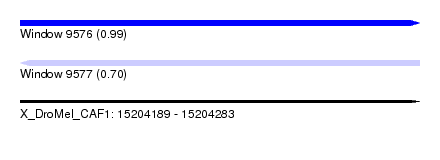
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,204,189 – 15,204,283 |
| Length | 94 |
| Max. P | 0.989493 |

| Location | 15,204,189 – 15,204,283 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -20.94 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15204189 94 + 22224390 UUAUGGGUUAUGGGUUAUGGGCUAUGGGCUAUGGGCUAUGGGCU----GCAUGUGC--AGCAUGUGCUGUAUGUGCCCUAUGUGCCUUAUGUGCCAGGAG .(((((.(((((...))))).)))))((((((((((((((((..----((((((((--(((....))))))))))))))))).))).)))).)))..... ( -38.20) >DroSec_CAF1 24767 80 + 1 UUAUGGGUUA--------------UGGGCUAUGGGUUAUUGGCU----GCAUGUGC--AGCAUGUGCUCUAUGUGCCCUAUGUGCCUCAUGUGCCAGGAG ...(((..((--------------(((((((((((......(((----((....))--)))..(..(.....)..))))))).))).))))..))).... ( -28.80) >DroSim_CAF1 23834 80 + 1 UUAUGGGUUA--------------UGGGCUAUGGGUUAUUGGCC----GCAUGUGC--AGCAUGUGCUCUAUGUGCCCUAUGUGCCUCAUGUUCUAGGAG ...(((..((--------------((((((((((((...(((.(----(((((...--..))))))..)))...)))).))).))).))))..))).... ( -26.50) >DroYak_CAF1 24218 100 + 1 UUAUGGGUUCUCGGUUAUGGGUUAUGGGCUAUGGGCUAUGGACUAUGAGCAUGUGCUCGGUGUGUGCCCUAUGUGCCCUAUGUGCUUCAUGUGCCAGGAG ........((..((((((((((...(((((((((((.(((.(((..((((....))))))).))))))).))).)))).....)).))))).)))..)). ( -34.00) >consensus UUAUGGGUUA______________UGGGCUAUGGGCUAUGGGCU____GCAUGUGC__AGCAUGUGCUCUAUGUGCCCUAUGUGCCUCAUGUGCCAGGAG ..........................((((((((((...((((.....(((..(((...)))..))).......)))).....))).)))).)))..... (-20.94 = -20.75 + -0.19)


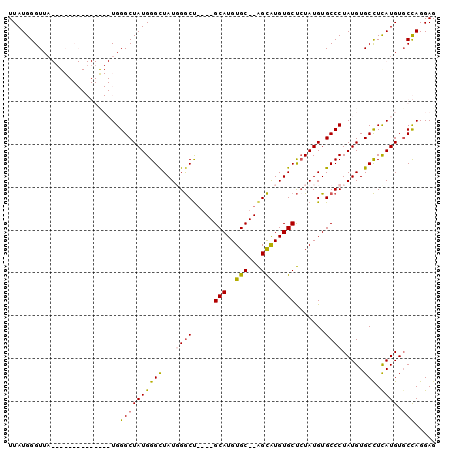
| Location | 15,204,189 – 15,204,283 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -10.82 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15204189 94 - 22224390 CUCCUGGCACAUAAGGCACAUAGGGCACAUACAGCACAUGCU--GCACAUGC----AGCCCAUAGCCCAUAGCCCAUAGCCCAUAACCCAUAACCCAUAA ....(((.......(((.....((((.......((....(((--((....))----))).....)).....))))...)))......))).......... ( -19.72) >DroSec_CAF1 24767 80 - 1 CUCCUGGCACAUGAGGCACAUAGGGCACAUAGAGCACAUGCU--GCACAUGC----AGCCAAUAACCCAUAGCCCA--------------UAACCCAUAA ....(((...(((.(((.....((((.......))....(((--((....))----)))......))....)))))--------------)...)))... ( -18.60) >DroSim_CAF1 23834 80 - 1 CUCCUAGAACAUGAGGCACAUAGGGCACAUAGAGCACAUGCU--GCACAUGC----GGCCAAUAACCCAUAGCCCA--------------UAACCCAUAA ..........(((.((......((((.............(((--((....))----)))............)))).--------------...))))).. ( -15.51) >DroYak_CAF1 24218 100 - 1 CUCCUGGCACAUGAAGCACAUAGGGCACAUAGGGCACACACCGAGCACAUGCUCAUAGUCCAUAGCCCAUAGCCCAUAACCCAUAACCGAGAACCCAUAA (((.((((.......)).))..((((.....((((.......((((....)))).((.....))))))...)))).............)))......... ( -22.30) >consensus CUCCUGGCACAUGAGGCACAUAGGGCACAUAGAGCACAUGCU__GCACAUGC____AGCCAAUAACCCAUAGCCCA______________UAACCCAUAA ......................((((......(((....)))((((....)))).................))))......................... (-10.82 = -11.07 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:08 2006