
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,187,206 – 15,187,377 |
| Length | 171 |
| Max. P | 0.938912 |

| Location | 15,187,206 – 15,187,326 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -34.91 |
| Energy contribution | -35.33 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
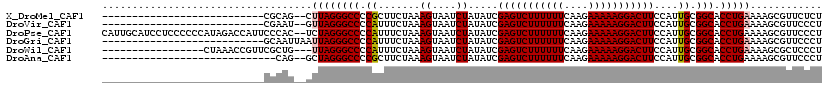
>X_DroMel_CAF1 15187206 120 - 22224390 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCUCUGGAAGCCUACUUCGCCGUCCAGCCGAGGCCAUCCGGUGAGAAAAUCGCAGCGAUUGCCGAAAAGCUGGAUUUGAAGAAAA .....(((.((((((....((.((((....)).)).)).))))))))).(((((..(((((((...(((.((((.((((.....)))).).))).))).....))))))).))))).... ( -40.90) >DroVir_CAF1 10918 120 - 1 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCUGGAAGCCUAUUUCGCCGUCCAGCCGAGGCCGUCCGGUGAGAAAAUCGCUGCCAUUGCCGAAAAGCUGGACUUGAAGAAAA ((((.(((.((((((..(.((.((((....)).)).)))))))))))).))))...(((((((...(((....((((((.....)))))).....))).....))))))).......... ( -43.40) >DroPse_CAF1 10338 120 - 1 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCUGGAAGCCUACUUUGCCGUCCAGCCGAGGCCAUCCGGUGAGAAAAUCGCGGCCAUCGCAGAAAAGCUGGACUUGAAGAAAA .....(((.((((((..(.((.((((....)).)).)))))))))))).((((...((((((((((((((...((((......)))).)))).))).......)))))))..)))).... ( -41.31) >DroGri_CAF1 11634 120 - 1 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCUGGAAGCCUACUUCGCCGUCCAGCCGAGGCCGUCGGGUGAGAAAAUCGCUGCCAUUGCCGAAAAGCUGGAUUUGAAGAAAA .....(((.((((((..(.((.((((....)).)).)))))))))))).(((((..(((((((...(((.((.((((((.....))))..)))).))).....))))))).))))).... ( -40.60) >DroWil_CAF1 13335 120 - 1 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGCUCCCUGGAAGCCUACUUUGCCGUCCAGCCGAGACCAUCUGGGGAGAAAAUCGCCGCCAUUGCCGAAAAGCUGGAUUUGAAGAAAA .....(((.((((((..(.(((..((....)).))).).))))))))).((((...(((((((..........(((((.(.....).)).)))..........)))))))..)))).... ( -32.65) >DroAna_CAF1 11634 120 - 1 GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCUGGAAGCCUACUUCGCCGUCCAGCCGAGACCAUCCGGUGAGAAAAUCGCUGCCAUUGCCGAAAAGCUGGAUUUGAAGAAAA .....(((.((((((..(.((.((((....)).)).)))))))))))).(((((..(((((((((.(......((((((.....))))))......)))....))))))).))))).... ( -37.20) >consensus GAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCUGGAAGCCUACUUCGCCGUCCAGCCGAGGCCAUCCGGUGAGAAAAUCGCUGCCAUUGCCGAAAAGCUGGAUUUGAAGAAAA .....(((.((((((..(.((.((((....)).)).)))))))))))).(((((..(((((((...(((....((((((.....)))))).....))).....))))))).))))).... (-34.91 = -35.33 + 0.42)



| Location | 15,187,286 – 15,187,377 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15187286 91 - 22224390 ---------------------------CGCAG--CUUAGGGCCCCGCUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCUCU ---------------------------(((..--.(((((...((((......((....)).....(((((((((((....))))))))))).....))))..)))))...)))...... ( -24.20) >DroVir_CAF1 10998 91 - 1 ---------------------------CGAAU--GUUAGGGCCCCAUUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCU ---------------------------.((((--(((((((((.((.......((....)).....(((((((((((....)))))))))))....)).))).))))....))))))... ( -23.30) >DroPse_CAF1 10418 118 - 1 CAUUGCAUCCUCCCCCCAUAGACCAUUCCCAC--UCUAGGGCCCCAUUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCU ....(((.......(((.((((..........--))))))).........................(((((((((((....)))))))))))....)))((.((((....)).)).)).. ( -22.20) >DroGri_CAF1 11714 93 - 1 ---------------------------GCAAUUAAUUAGGGCCCCAUUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCU ---------------------------(((((...((((((......)))))).............(((((((((((....)))))))))))..)))))((.((((....)).)).)).. ( -20.40) >DroWil_CAF1 13415 100 - 1 -----------------CUAAACCGUUCGCUG---UUAGGGCCCCAUUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGCUCCCU -----------------.......(..((((.---((((((((.((.......((....)).....(((((((((((....)))))))))))....)).))).)))))..))))..)... ( -25.00) >DroAna_CAF1 11714 89 - 1 -----------------------------CAG--GCUAGGGCCCCGCUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCU -----------------------------...--(((.(((..((((......((....)).....(((((((((((....))))))))))).....))))..)))....)))....... ( -22.00) >consensus ___________________________CGCAG__GUUAGGGCCCCAUUUCUAAAGUAAUCUAUAUCGAGUCUUUUUUCAAGAAAAAGGACUUCCAUUGCGGCACCUGAAAAGCGUUCCCU ...................................((((((((.((.......((....)).....(((((((((((....)))))))))))....)).))).)))))............ (-18.82 = -18.93 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:05 2006