
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,178,546 – 15,178,670 |
| Length | 124 |
| Max. P | 0.994927 |

| Location | 15,178,546 – 15,178,643 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -21.95 |
| Energy contribution | -23.04 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15178546 97 + 22224390 UUCAAAGUUAGAGGCGGAAAAACAUUUUAGGCCAACAU---------CGAUGUCUUCGAAAUAUCAGAAACAUCGAUGUU-UUCGGUGAAUCAGCGAUGUUUUUUCA ...............(((((((((((....((((((((---------((((((.(((.........))))))))))))))-...)))........))))))))))). ( -26.20) >DroSec_CAF1 6546 106 + 1 UUCGAAGCUAAAGGCGGAGAAACAUU-GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGAAAUCAUCGAUGAUUUGCCA ......((....((((..((....))-..))))..(((((((((((((((((.((..((....))..)).))))))))))....((....)))))))))....)).. ( -34.80) >DroSim_CAF1 6328 106 + 1 UUCGAAGCUAAAGGCGGAGAAACAUU-GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGGAAUCAUCGAUGAUUUGCCA ((((((......((((..((....))-..))))((((((((((((...(((...........)))...)))))))))))).))))))((((((....)))))).... ( -34.10) >consensus UUCGAAGCUAAAGGCGGAGAAACAUU_GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGGAAUCAUCGAUGAUUUGCCA ((((.((.....((((.............)))).(((((((.....))))))))).)))).......(((((((((((.............)))))))))))..... (-21.95 = -23.04 + 1.09)

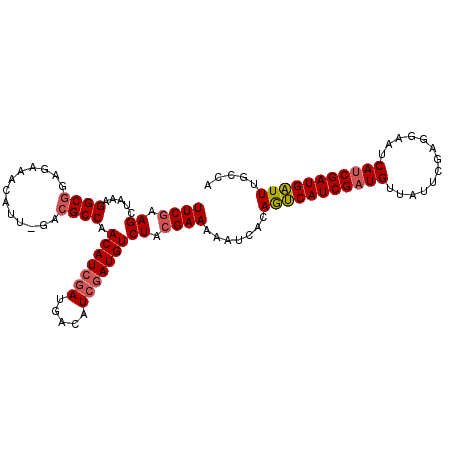

| Location | 15,178,546 – 15,178,643 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15178546 97 - 22224390 UGAAAAAACAUCGCUGAUUCACCGAA-AACAUCGAUGUUUCUGAUAUUUCGAAGACAUCG---------AUGUUGGCCUAAAAUGUUUUUCCGCCUCUAACUUUGAA .(.((((((((((.((...)).))).-(((((((((((((.(((....))).))))))))---------))))).........))))))).)............... ( -23.90) >DroSec_CAF1 6546 106 - 1 UGGCAAAUCAUCGAUGAUUUCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC-AAUGUUUCUCCGCCUUUAGCUUCGAA .(((....(((((((((............((((((((.((((((....)))))).)))))))))))))))))..((((..-..........))))....)))..... ( -32.20) >DroSim_CAF1 6328 106 - 1 UGGCAAAUCAUCGAUGAUUCCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC-AAUGUUUCUCCGCCUUUAGCUUCGAA .(((....((((((((((((...)))).(((((((((.((((((....)))))).)))))))))))))))))..((((..-..........))))....)))..... ( -33.40) >consensus UGGCAAAUCAUCGAUGAUUCCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC_AAUGUUUCUCCGCCUUUAGCUUCGAA .......(((((((((((((...))))..))))))))).........((((..((((((((.....))))))))((((.............))))........)))) (-19.65 = -20.52 + 0.87)

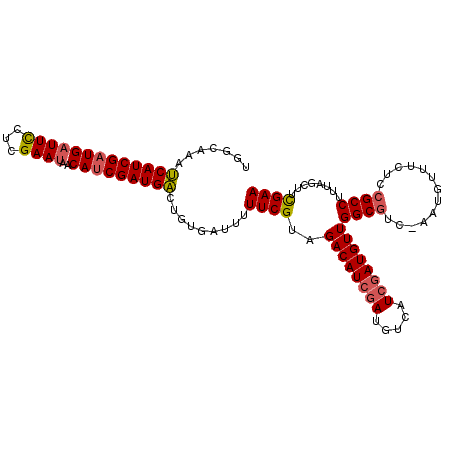
| Location | 15,178,573 – 15,178,670 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15178573 97 + 22224390 UAGGCCAACAU---------CGAUGUCUUCGAAAUAUCAGAAACAUCGAUGUU-UUCGGUGAAUCAGCGAUGUUUUUUCAUCUCUAUUGCUCAUCGGAGAAUAAAAG ......(((((---------((((((.(((.........))))))))))))))-((((((((...((.((((......)))).)).....))))))))......... ( -27.20) >DroSec_CAF1 6572 107 + 1 GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGAAAUCAUCGAUGAUUUGCCAUCUCUAUUGCGCAUCGGAGGAAAAAAG ..(.((..(((((((((((((((((.((..((....))..)).))))))))))....((....)))))))))...(((((.......)).)))..)).)........ ( -31.10) >DroSim_CAF1 6354 106 + 1 GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGGAAUCAUCGAUGAUUUGCCAUCUCUAUUGCGCAUCGGAGGAA-AAAG ....((((((((((((((...(((...........)))...)))))))))))).(((((.((((((....))))))((((.......)).)).)))))))..-.... ( -29.20) >consensus GACGCCAACAUCGAUGACAUCGAUGUCUACGAAAAAUCACAGUCAUCGAUGUUAUUCGAGGAAUCAUCGAUGAUUUGCCAUCUCUAUUGCGCAUCGGAGGAAAAAAG .......(((((((.....)))))))..............(((((((((((.............))))))))))).....(((((..(....)..)))))....... (-18.75 = -18.95 + 0.20)


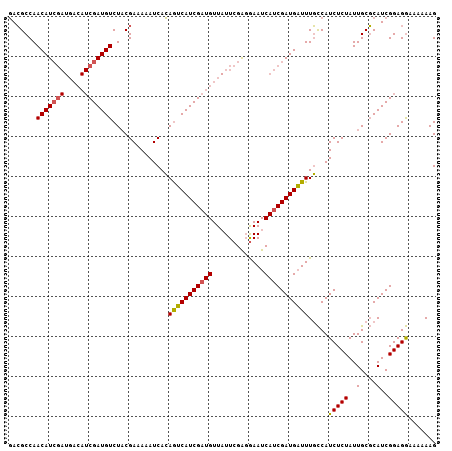
| Location | 15,178,573 – 15,178,670 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15178573 97 - 22224390 CUUUUAUUCUCCGAUGAGCAAUAGAGAUGAAAAAACAUCGCUGAUUCACCGAA-AACAUCGAUGUUUCUGAUAUUUCGAAGACAUCG---------AUGUUGGCCUA ...........((.((((...(((.((((......)))).))).)))).))..-(((((((((((((.(((....))).))))))))---------)))))...... ( -27.10) >DroSec_CAF1 6572 107 - 1 CUUUUUUCCUCCGAUGCGCAAUAGAGAUGGCAAAUCAUCGAUGAUUUCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC ............(((((.(((((..((((((......((((.(....))))).....(((((((.((((((....)))))).)))))))))))))..)))))))))) ( -35.60) >DroSim_CAF1 6354 106 - 1 CUUU-UUCCUCCGAUGCGCAAUAGAGAUGGCAAAUCAUCGAUGAUUCCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC ....-.......(((((.(((((..(((((((..(((((((((((((...))))..)))))))))((((((....)))))).......)))))))..)))))))))) ( -36.10) >consensus CUUUUUUCCUCCGAUGCGCAAUAGAGAUGGCAAAUCAUCGAUGAUUCCUCGAAUAACAUCGAUGACUGUGAUUUUUCGUAGACAUCGAUGUCAUCGAUGUUGGCGUC ...............((((...((((((.((...(((((((((((((...))))..)))))))))..)).))))))....((((((((.....)))))))).)))). (-21.30 = -22.83 + 1.53)


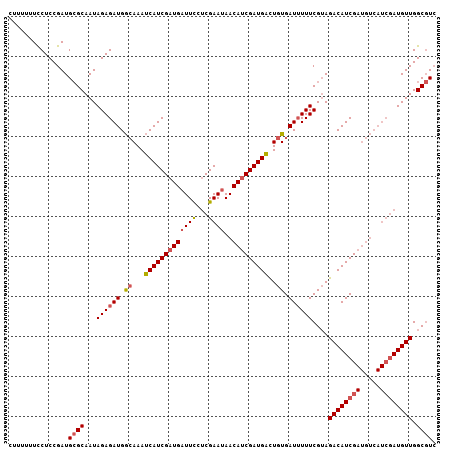
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:02 2006