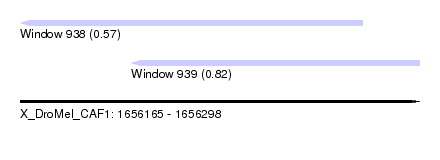
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,656,165 – 1,656,298 |
| Length | 133 |
| Max. P | 0.822823 |

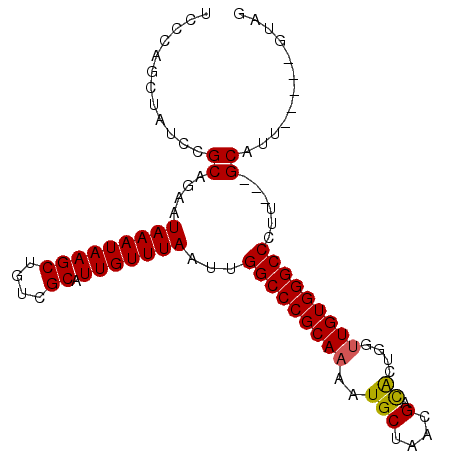
| Location | 1,656,165 – 1,656,279 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.55 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.39 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1656165 114 - 22224390 AAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCAAACGACACUGGUUGUGGGCCCUU---GCAUUGUAUUGUAGAUGGCAAUCAUAAAAUCAUAAAUGUCGGACAAUCGCU--- .....(((....(((.(((......(((((((.....))..(((((....))))))))))...---))).)))(((((...(((((................))))).))))).)))--- ( -27.19) >DroPse_CAF1 41237 97 - 1 GCAUAAGCUGUCAGAUUGUUUUAACGAGCAGCG-----------GGGCAGGUUCAAGUUCAAUUCUGGAAU----AGUAGAUUCCAGCUG--------AAAAUAUCGGUCAAUGUACAAA ((((..(((((....((((....))))))))).-----------.(((.(((.....((((...(((((((----.....))))))).))--------))...))).))).))))..... ( -23.00) >DroSec_CAF1 21023 109 - 1 AAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCUAACGACGCUGGUUGUGGGCCCUU---GCAUU-----GUAGAUGGCAAUCAUAAAAUCAUAAAUGCCGGACAAUCGCU--- ((((((((....)).))))))....(((((((((...((.......))...)))))))))...---(((((-----((...(((((................))))).))))).)).--- ( -32.59) >DroSim_CAF1 10281 109 - 1 AAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCUAACGACGCUGGUUGUGGGCCCUU---GCAUU-----GUAGAUGGCAAUCAUAAAAUCAUAAAUGCCGGACAAUCGCU--- ((((((((....)).))))))....(((((((((...((.......))...)))))))))...---(((((-----((...(((((................))))).))))).)).--- ( -32.59) >DroYak_CAF1 21613 104 - 1 AAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCA-AAUGCUAACGAUAGUGGCUGUGGGCCCUUCCUGCAUU----UGUAGAUGGCAAUCA--------AAAAUGUCGGACAAUCGCU--- ............((((((.......((((((((-..(((((....)))))..))))))))..(((.(((((----(...(((....))).--------.)))))).))))))).)).--- ( -32.50) >DroPer_CAF1 43169 97 - 1 GCAUAAGCUGUCAGAUUGUUUUAACGAGCAGCG-----------GGGCAGGUUCAAGUUCAAUUCUGGAAU----AGUAGAUUCCAGCUG--------AAAAUAUCGGUCAAUGUACAAA ((((..(((((....((((....))))))))).-----------.(((.(((.....((((...(((((((----.....))))))).))--------))...))).))).))))..... ( -23.00) >consensus AAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCA_AAUGCUAACGACGCUGGUUGUGGGCCCUU___GCAUU_____GUAGAUGGCAAUCA________AAAAUGUCGGACAAUCGCU___ ............(((((((((....((((((((...................))))))))......((...............)).....................))))))).)).... (-12.72 = -12.39 + -0.33)

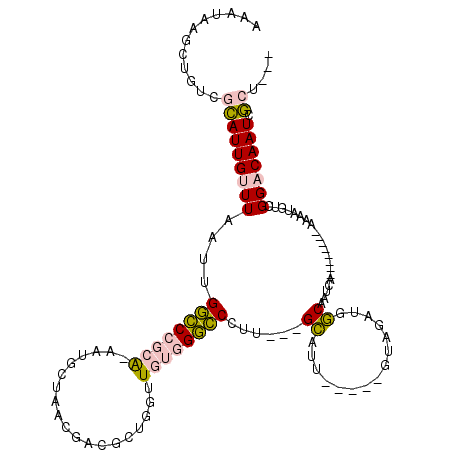
| Location | 1,656,202 – 1,656,298 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.54 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1656202 96 - 22224390 UCCCAGCUAUCCGCAGAAUAAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCAAACGACACUGGUUGUGGGCCCUU---GCAUUGUAUUGUAG ............((((..(((((((((....)).)))))))...(((((((.....))..(((((....)))))))))).))---))............ ( -24.60) >DroSec_CAF1 21060 91 - 1 UCCCAGCUAUCCGCAGAAUAAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCUAACGACGCUGGUUGUGGGCCCUU---GCAUU-----GUAG ............((((..(((((((((....)).)))))))...(((((((((...((.......))...))))))))).))---))...-----.... ( -27.40) >DroSim_CAF1 10318 91 - 1 UCCCAGCUAUCCGCAGAAUAAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCUAACGACGCUGGUUGUGGGCCCUU---GCAUU-----GUAG ............((((..(((((((((....)).)))))))...(((((((((...((.......))...))))))))).))---))...-----.... ( -27.40) >DroYak_CAF1 21642 83 - 1 -----------CGCAGAAUAAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCA-AAUGCUAACGAUAGUGGCUGUGGGCCCUUCCUGCAUU----UGUAG -----------.((((..(((((((((....)).)))))))...((((((((-..(((((....)))))..))))))))....))))...----..... ( -29.90) >consensus UCCCAGCUAUCCGCAGAAUAAAUAAGCUGUCGCAUUGUUUAAUUGGCCCGCAAAAUGCUAACGACACUGGUUGUGGGCCCUU___GCAUU_____GUAG ............((....(((((((((....)).)))))))...(((((((((..(((....).))....)))))))))......))............ (-22.79 = -22.60 + -0.19)

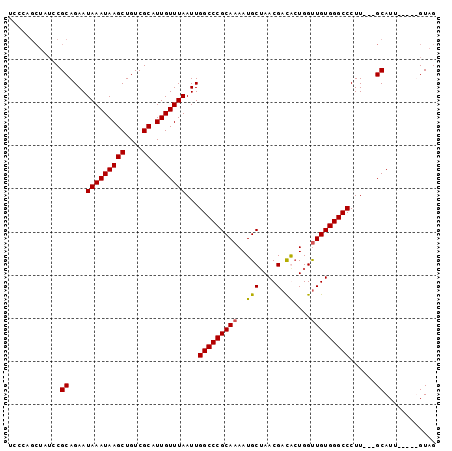
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:39 2006