
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,147,566 – 15,147,707 |
| Length | 141 |
| Max. P | 0.996379 |

| Location | 15,147,566 – 15,147,667 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -27.04 |
| Energy contribution | -28.35 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15147566 101 + 22224390 UGCAACUGUUUGCAUUGCUUUUGCCAAUGCAAUUGCAAUUGCAAUUGCAAUUGCUCCUGCACUUUGUUUUGCACUCGCCAAUGUGUGU----GUGUUUUUUGCGA ((((.....(((((((((....).))))))))..((((((((....))))))))...))))..((((...((((.(((......))).----)))).....)))) ( -31.60) >DroSec_CAF1 58588 105 + 1 UGCAACUGUUUGCAUUGCUUUUGCCAAUGCAAUUGCCAUUGCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGUGUGUGUGUAUUUUGCGA (((((....))))).(((....((...(((((((((....)))))))))...))....)))..((((..(((((.(((((.....)).))).)))))....)))) ( -32.20) >DroSim_CAF1 50056 105 + 1 UGCAACUGUUUGCAUUGCUUUUGCCAAUGCAAUUGCCAUUGCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGUGUGUGUGUAUUUUGCGA (((((....))))).(((....((...(((((((((....)))))))))...))....)))..((((..(((((.(((((.....)).))).)))))....)))) ( -32.20) >DroEre_CAF1 65108 101 + 1 UGCAACUGUUUGCAUUGCUUUUGCCAAUGCAAUGGCAAUUGCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUCUGU----GUGUAUUUUUCGA ((((.....(((((((((....).)))))))).(((((((((....)))))))))..))))..(((...(((((.((........)).----))))).....))) ( -30.30) >consensus UGCAACUGUUUGCAUUGCUUUUGCCAAUGCAAUUGCAAUUGCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGU____GUGUAUUUUGCGA ((((.....(((((((((....).))))))))..((((((((....))))))))...))))..((((..(((((.(((........)))...)))))....)))) (-27.04 = -28.35 + 1.31)



| Location | 15,147,566 – 15,147,667 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.41 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -26.83 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15147566 101 - 22224390 UCGCAAAAAACAC----ACACACAUUGGCGAGUGCAAAACAAAGUGCAGGAGCAAUUGCAAUUGCAAUUGCAAUUGCAUUGGCAAAAGCAAUGCAAACAGUUGCA ..((((.......----.....(((((.(...(((.......(((((((..((((((((....))))))))..))))))).)))...)))))).......)))). ( -29.61) >DroSec_CAF1 58588 105 - 1 UCGCAAAAUACACACACACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGCAAUGGCAAUUGCAUUGGCAAAAGCAAUGCAAACAGUUGCA ((((.......................)))).((((........))))...((...(((((((((....)))))))))...))....(((((.......))))). ( -26.10) >DroSim_CAF1 50056 105 - 1 UCGCAAAAUACACACACACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGCAAUGGCAAUUGCAUUGGCAAAAGCAAUGCAAACAGUUGCA ((((.......................)))).((((........))))...((...(((((((((....)))))))))...))....(((((.......))))). ( -26.10) >DroEre_CAF1 65108 101 - 1 UCGAAAAAUACAC----ACAGACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGCAAUUGCCAUUGCAUUGGCAAAAGCAAUGCAAACAGUUGCA .............----..........((...(((.......(((((((..((((((((....))))))))..))))))).)))...))..(((((....))))) ( -28.90) >consensus UCGCAAAAUACAC____ACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGCAAUGGCAAUUGCAUUGGCAAAAGCAAUGCAAACAGUUGCA ...........................((...(((.......(((((((..((((((((....))))))))..))))))).)))...))..(((((....))))) (-26.83 = -27.33 + 0.50)

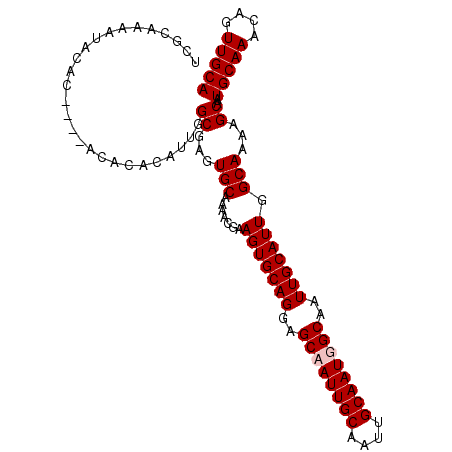

| Location | 15,147,606 – 15,147,707 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15147606 101 + 22224390 GCAAUUGCAAUUGCUCCUGCACUUUGUUUUGCACUCGCCAAUGUGUGU----GUGUUUUUUGCGAACUUGUUUUGCAAUUGCGUUUUAAAAUAAAUUACAUCAGU ((((((((((.(((....))).(((((...((((.(((......))).----)))).....)))))......))))))))))....................... ( -25.70) >DroSec_CAF1 58628 101 + 1 GCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGUGUGUGUGUAUUUUGCGAACUUGUUUUGCAAUUGCGUUUUAAAAUAAAUUACAU---- ((((((((((.(((....))).(((((..(((((.(((((.....)).))).)))))....)))))......))))))))))...................---- ( -30.00) >DroSim_CAF1 50096 105 + 1 GCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGUGUGUGUGUAUUUUGCGAACUUGUUUUGCAAUUGCGUUUUAAAAUAAAUUACAUCAGU ((((((((((.(((....))).(((((..(((((.(((((.....)).))).)))))....)))))......))))))))))....................... ( -30.00) >DroEre_CAF1 65148 101 + 1 GCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUCUGU----GUGUAUUUUUCGAACUUGUUUUGCAAUUGCGUUUUAAAAUAAAUUACAUCAGU ((((((((((.(((....))).((((...(((((.((........)).----))))).....))))......))))))))))....................... ( -23.70) >consensus GCAAUUGCAAUUGCUCCUGCACUUCGUUUUGCACUCGCCAAUGUGUGU____GUGUAUUUUGCGAACUUGUUUUGCAAUUGCGUUUUAAAAUAAAUUACAUCAGU ((((((((((.(((....))).(((((..(((((.(((........)))...)))))....)))))......))))))))))....................... (-23.14 = -23.95 + 0.81)

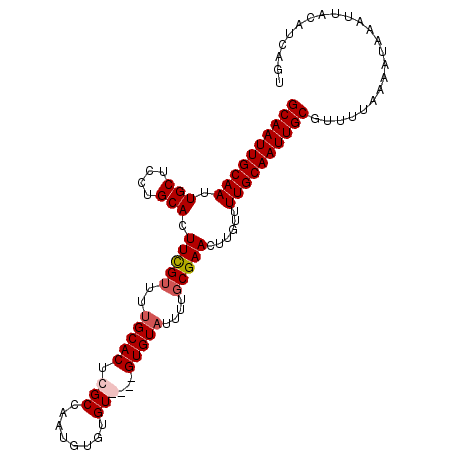
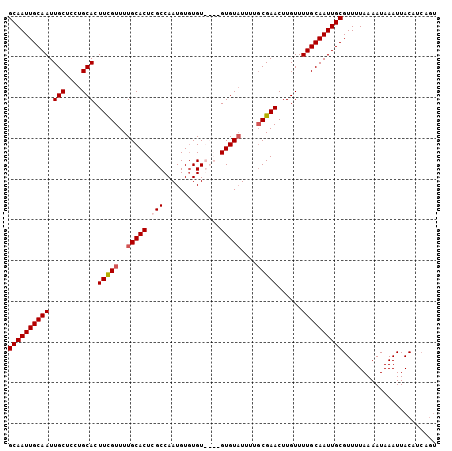
| Location | 15,147,606 – 15,147,707 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15147606 101 - 22224390 ACUGAUGUAAUUUAUUUUAAAACGCAAUUGCAAAACAAGUUCGCAAAAAACAC----ACACACAUUGGCGAGUGCAAAACAAAGUGCAGGAGCAAUUGCAAUUGC .......................((((((((........(((((.........----..........)))))((((........))))...))))))))...... ( -19.31) >DroSec_CAF1 58628 101 - 1 ----AUGUAAUUUAUUUUAAAACGCAAUUGCAAAACAAGUUCGCAAAAUACACACACACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGC ----...................((((((((........(((((.......................)))))((((........))))...))))))))...... ( -19.10) >DroSim_CAF1 50096 105 - 1 ACUGAUGUAAUUUAUUUUAAAACGCAAUUGCAAAACAAGUUCGCAAAAUACACACACACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGC .......................((((((((........(((((.......................)))))((((........))))...))))))))...... ( -19.10) >DroEre_CAF1 65148 101 - 1 ACUGAUGUAAUUUAUUUUAAAACGCAAUUGCAAAACAAGUUCGAAAAAUACAC----ACAGACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGC .......................((((((((((..(((..((...........----...))..)))((...((((........))))...))..)))))))))) ( -19.34) >consensus ACUGAUGUAAUUUAUUUUAAAACGCAAUUGCAAAACAAGUUCGCAAAAUACAC____ACACACAUUGGCGAGUGCAAAACGAAGUGCAGGAGCAAUUGCAAUUGC .......................((((((((........(((((.......................)))))((((........))))...))))))))...... (-17.65 = -17.90 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:45 2006