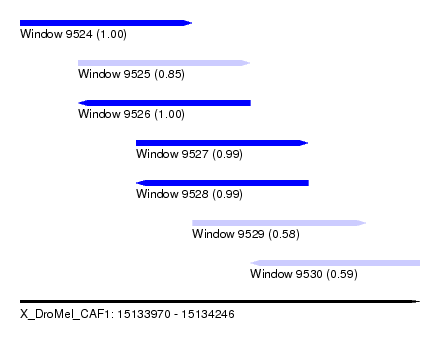
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,133,970 – 15,134,246 |
| Length | 276 |
| Max. P | 0.999452 |

| Location | 15,133,970 – 15,134,089 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15133970 119 + 22224390 CUACGAGUACGAGCAUAUGUGCUGAAAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGAGUUGGGAAAAGUUUCGAUUUACGAAAA-AC ...((.(((((.((.....(((((....))))))).)))))(((((....)))))(((((((((.......((((((((.......))))))))......)))))))))..))....-.. ( -39.52) >DroSec_CAF1 45418 119 + 1 CUGCGAGUACGAGCAUAUGUGCUGAUAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-AC .((((.(((((.((.....(((((....))))))).))))).))))((((((...(((((((((((((((((((.........)))))..)))))).....))))))))..))))))-.. ( -39.30) >DroSim_CAF1 36158 119 + 1 CUACGAGUACGAGCAUAUGUGCUGAAAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-AC ...((.(((((.((.....(((((....))))))).)))))(((((....)))))(((((((((((((((((((.........)))))..)))))).....))))))))..))....-.. ( -39.30) >DroEre_CAF1 52345 108 + 1 AU------------UUAUGUGCUGAAAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCACUUUUUUGUCGACUUGGGAAAAGUUUCGAUUUACGAAAAAAA ..------------.(((((((((.......)))))))))((((((....))))))(((((((((((((..((((..........)))).)))))).....)))))))............ ( -35.80) >DroYak_CAF1 53073 107 + 1 AU------------AUAUGUGCUGAAAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAAGGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-AC ..------------.(((((((((.......)))))))))((((((....))))))(((((((((((((.......(((....)))....)))))).....))))))).........-.. ( -32.60) >consensus CU_CGAGUACGAGCAUAUGUGCUGAAAUCAGCGGCGCGUGCGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA_AC ...............(((((((((.......)))))))))((((((....))))))((((((((((((((((((.........)))))..)))))).....)))))))............ (-32.96 = -33.56 + 0.60)

| Location | 15,134,010 – 15,134,129 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134010 119 + 22224390 CGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGAGUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUUCGAAAAUCUGCU ((((((....)))))).(((((....((..((((....(((((((((.((((((..(....)..)))))))))))))-)).....(((.....))).))))..)).)))))......... ( -32.50) >DroSec_CAF1 45458 119 + 1 CGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCU ((((((....)))))).(((.(((..((((((......)))))).)))(((.((((((...(((((.....))))).-...))))))..)))................)))......... ( -31.00) >DroSim_CAF1 36198 119 + 1 CGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCU ((((((....)))))).(((.(((..((((((......)))))).)))(((.((((((...(((((.....))))).-...))))))..)))................)))......... ( -31.00) >DroEre_CAF1 52373 120 + 1 CGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCACUUUUUUGUCGACUUGGGAAAAGUUUCGAUUUACGAAAAAAAAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCU ((((((....)))))).(((........(((((((..........)))(((.((((((...(((((.....))))).....))))))..)))...)))).........)))......... ( -28.63) >DroYak_CAF1 53101 119 + 1 CGCGCAUUUUUGCGCGAUCGAAACCCGAGAAGGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCU ((((((....)))))).(((.....)))........(((.((((((..(((.((((((...(((((.....))))).-...))))))..))).(((.....)))....)))))).))).. ( -31.00) >consensus CGCGCAUUUUUGCGCGAUCGAAACCCGAGAACGAUUCAGUUUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA_ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCU ((((((....)))))).(((.....)))........(((.((((((..(((.((((((...(((((.....))))).....))))))..))).(((.....)))....)))))).))).. (-28.34 = -28.74 + 0.40)


| Location | 15,134,010 – 15,134,129 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.65 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134010 119 - 22224390 AGCAGAUUUUCGAAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAACUCAACAAAAAACUGAAUCGUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((..(((((.((((((........((...((((((...-.(((((.....)))))...))))))..))..((......))..)))))))))))..))...((((((....)))))) ( -32.60) >DroSec_CAF1 45458 119 - 1 AGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAAACUGAAUCGUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((((.((.((((((((........(((..((((((...-.(((((.....)))))...)))))).)))..((......))..)))))))).))....))))(((((....))))). ( -35.80) >DroSim_CAF1 36198 119 - 1 AGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAAACUGAAUCGUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((((.((.((((((((........(((..((((((...-.(((((.....)))))...)))))).)))..((......))..)))))))).))....))))(((((....))))). ( -35.80) >DroEre_CAF1 52373 120 - 1 AGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUUUUUUUUCGUAAAUCGAAACUUUUCCCAAGUCGACAAAAAAGUGAAUCGUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((((.((.((((((((........(((..((((((.....(((((.....)))))...)))))).))).((......))...)))))))).))....))))(((((....))))). ( -35.30) >DroYak_CAF1 53101 119 - 1 AGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAAACUGAAUCCUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((..((((((((((..........(((..((((((...-.(((((.....)))))...)))))).)))..((......)).)))).)).))))..))...((((((....)))))) ( -33.50) >consensus AGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU_UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAAACUGAAUCGUUCUCGGGUUUCGAUCGCGCAAAAAUGCGCG ....((..((((.(((((((........(((..((((((.....(((((.....)))))...)))))).)))..((......))..)))))))))))..))...((((((....)))))) (-32.34 = -32.74 + 0.40)


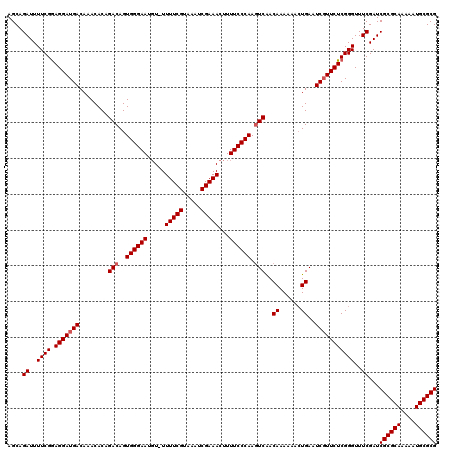
| Location | 15,134,050 – 15,134,169 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -23.72 |
| Energy contribution | -25.56 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134050 119 + 22224390 UUUUUGUUGAGUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUUCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAUCCACAAACACAUAC ....(((((.((((((.....((((((.....((.((-((((...........)))))).)).....)))))).....))))))))))).....(((((((........))))))).... ( -29.60) >DroSec_CAF1 45498 119 + 1 UUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUAC ........(((.((((((...(((((.....))))).-...))))))..)))(((((((((..((....))......(((..((.(((((....))))).))..))).)))))))))... ( -31.10) >DroSim_CAF1 36238 119 + 1 UUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUAC ........(((.((((((...(((((.....))))).-...))))))..)))(((((((((..((....))......(((..((.(((((....))))).))..))).)))))))))... ( -31.10) >DroEre_CAF1 52413 118 + 1 UUUUUGUCGACUUGGGAAAAGUUUCGAUUUACGAAAAAAAAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAUCCAACACUAACG--AUCGCCCAGCCACAAACACAUAG ........(((.((((((...(((((.....))))).....))))))..)))(((((((((.......(((.....((......))...(....)--.))).......)))))))))... ( -23.84) >DroYak_CAF1 53141 117 + 1 UUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA-ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACG--UUUGCCCAGCCACAAACACAUAG ........(((.((((((...(((((.....))))).-...))))))..)))(((((((((..((....))......(((..((.(((......)--)).))..))).)))))))))... ( -29.20) >consensus UUUUUGUUGACUUGGGAAAAGUUUCGAUUUACGAAAA_ACAUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUAC ........(((.((((((...(((((.....))))).....))))))..)))(((((((((..((....))......(((..((.(((((....))))).))..))).)))))))))... (-23.72 = -25.56 + 1.84)

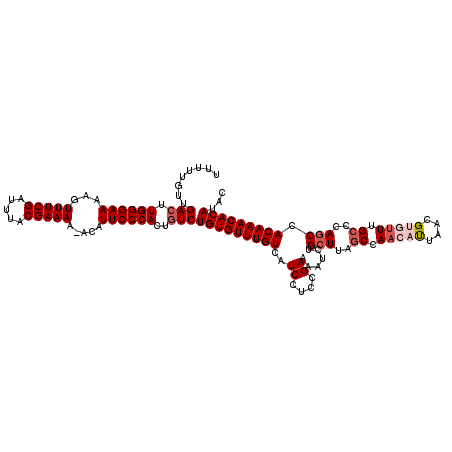
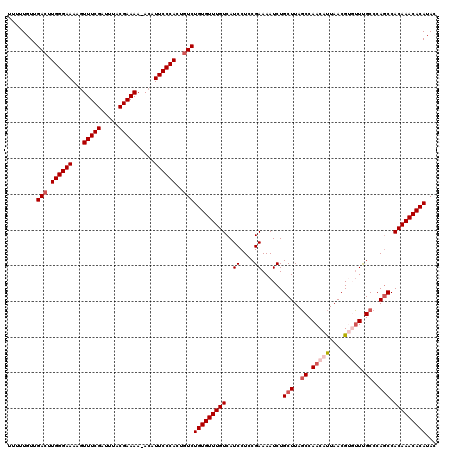
| Location | 15,134,050 – 15,134,169 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -28.52 |
| Energy contribution | -30.12 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134050 119 - 22224390 GUAUGUGUUUGUGGAUGGGCAAACACGUUAAUGUUGGCUAAGCAGAUUUUCGAAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAACUCAACAAAAA ((.(((((((((.(...(((.((((......)))).)))...)......((....))..))))))))).))..((((((...-.(((((.....)))))...))))))............ ( -31.20) >DroSec_CAF1 45498 119 - 1 GUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUAAGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAA ...(((((((((.(((.(((.((((......)))).))).)))......((....))..)))))))))(((..((((((...-.(((((.....)))))...)))))).)))........ ( -35.00) >DroSim_CAF1 36238 119 - 1 GUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUAAGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAA ...(((((((((.(((.(((.((((......)))).))).)))......((....))..)))))))))(((..((((((...-.(((((.....)))))...)))))).)))........ ( -35.00) >DroEre_CAF1 52413 118 - 1 CUAUGUGUUUGUGGCUGGGCGAU--CGUUAGUGUUGGAUAAGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUUUUUUUUCGUAAAUCGAAACUUUUCCCAAGUCGACAAAAA ...(((((((((.(((...(((.--.(((..((((.....)))))))..)))..)).).)))))))))(((..((((((.....(((((.....)))))...)))))).)))........ ( -32.20) >DroYak_CAF1 53141 117 - 1 CUAUGUGUUUGUGGCUGGGCAAA--CGUUAAUGUUGGCUAAGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU-UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAA ...(((((((((.(((.(((.((--((....)))).))).)))......((....))..)))))))))(((..((((((...-.(((((.....)))))...)))))).)))........ ( -34.30) >consensus GUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUAAGCAGAUUUUCGGAGGAUGACAAACACAGACAGUGGGAAUGU_UUUUCGUAAAUCGAAACUUUUCCCAAGUCAACAAAAA ...(((((((((.(((.(((.((((......)))).))).)))......((....))..)))))))))(((..((((((.....(((((.....)))))...)))))).)))........ (-28.52 = -30.12 + 1.60)



| Location | 15,134,089 – 15,134,209 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -17.43 |
| Consensus MFE | -13.17 |
| Energy contribution | -15.01 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134089 120 + 22224390 AUUCCCACUGUCUGUGUUUGUCAUCCUUCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAUCCACAAACACAUACAAAUGCAUACAAAAAAAAAACUAUCACAUUCGCAUCGACA .........((((((((((((..((....))...........((.(((((....))))).))......))))))))).....((((.........................)))).))). ( -16.61) >DroSec_CAF1 45537 117 + 1 AUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUACAAAUGCAUACGAA---AAAACUAUCACAUUCUCAUCGACA ........(((.(((((((((..((....))......(((..((.(((((....))))).))..))).))))))))).)))............---........................ ( -19.10) >DroSim_CAF1 36277 117 + 1 AUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUACAAAUGCAUACGAA---AAAACUAUCACAUUCGCAUCGACA .........((((((((((((..((....))......(((..((.(((((....))))).))..))).))))))))).....((((.......---...............)))).))). ( -20.15) >DroEre_CAF1 52453 110 + 1 AUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAUCCAACACUAACG--AUCGCCCAGCCACAAACACAUAGAAAUGCC-----A---AAAACUAUCACAUUCGCAUCGACA .........((((((((((((.......(((.....((......))...(....)--.))).......))))))))).....((((.-----.---...............)))).))). ( -12.87) >DroYak_CAF1 53180 110 + 1 AUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACG--UUUGCCCAGCCACAAACACAUAGAAAUGCU-----A---AAAACUAUCACAUUCGCAUCGACA .........((((((((((((..((....))......(((..((.(((......)--)).))..))).))))))))).....((((.-----.---...............)))).))). ( -18.43) >consensus AUUCCCACUGUCUGUGUUUGUCAUCCUCCGAAAAUCUGCUUAGCCAACAUUAACGUGUUUGCCCAGCCACAAACACAUACAAAUGCAUAC_AA___AAAACUAUCACAUUCGCAUCGACA .........((((((((((((..((....))......(((..((.(((((....))))).))..))).))))))))).....((((.........................)))).))). (-13.17 = -15.01 + 1.84)

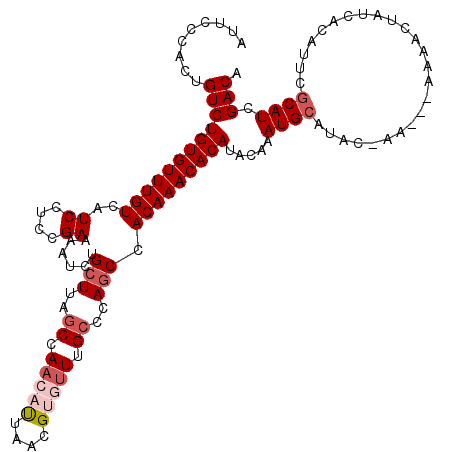
| Location | 15,134,129 – 15,134,246 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15134129 117 - 22224390 UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGCGAAUGUGAUAGUUUUUUUUUUGUAUGCAUUUGUAUGUGUUUGUGGAUGGGCAAACACGUUAAUGUUGGCUA ........................(((.(((((((.......(((((((((((((((........)))).)))))))))))((((((((......)))))))))))))))..))).. ( -29.50) >DroSec_CAF1 45577 114 - 1 UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGAGAAUGUGAUAGUUUU---UUCGUAUGCAUUUGUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUA ..(((((..((..(((((...(....(((((((((...)))))))))...).)))))..))---..)))))(((((...((((((((((......)))))))))).)))))...... ( -28.30) >DroSim_CAF1 36317 114 - 1 UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGCGAAUGUGAUAGUUUU---UUCGUAUGCAUUUGUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUA ........................(((.(((((((.......(((((((((((...(....---..)...)))))))))))((((((((......)))))))))))))))..))).. ( -28.00) >DroEre_CAF1 52493 107 - 1 UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGCGAAUGUGAUAGUUUU---U-----GGCAUUUCUAUGUGUUUGUGGCUGGGCGAU--CGUUAGUGUUGGAUA ....(..(((((.....((((((.(((.(((.((..(((((((.((..........))..)---)-----))))).))...))).....))).)))))).--.)))))..)...... ( -20.40) >DroYak_CAF1 53220 107 - 1 UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGCGAAUGUGAUAGUUUU---U-----AGCAUUUCUAUGUGUUUGUGGCUGGGCAAA--CGUUAAUGUUGGCUA ......((((((.((((((((((.(((..((((((...))))))(((((..(.((((....---.-----.......)))))..)))))))).)))))))--...))).)))))).. ( -23.00) >consensus UAAUAUGUUAAUUUUAUUUGUCUUGCCUCAUUGAUUGUGUCGAUGCGAAUGUGAUAGUUUU___UU_GUAUGCAUUUGUAUGUGUUUGUGGCUGGGCAAACACGUUAAUGUUGGCUA ....(..(((((....(((((((.(((.(((((((...))))))).((((((...................))))))............))).)))))))...)))))..)...... (-19.17 = -19.25 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:26 2006