
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,122,259 – 15,122,389 |
| Length | 130 |
| Max. P | 0.854054 |

| Location | 15,122,259 – 15,122,352 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -7.68 |
| Energy contribution | -9.44 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15122259 93 - 22224390 AAAAUAUAGAAAACGGAUUU-UGCAGUUUUGCCAUCAGAGCUGCUGGCUGUA--------------------CACGGCCAAAU-----AUAA-CAUUUUUUAGUAAUUACAUAUUUGCAC ......(((((((.(.....-.(((((((((....)))))))))((((((..--------------------..))))))...-----....-).)))))))((((........)))).. ( -24.20) >DroSec_CAF1 33914 97 - 1 AAAAUAUAGAAAACGGAUUU-UGCAGUUUUGCCAUCAGAGCUGC-GGCUGUA--------------------CACGGCCAAAUACUAUAUAA-AAUUUUUUAGUAAUUACAUAUUUGCAC ...((((((...........-.(((((((((....)))))))))-(((((..--------------------..))))).....))))))..-.........((((........)))).. ( -26.00) >DroEre_CAF1 40872 90 - 1 AAAAUAUAGAAAACGGAUUU-CGCAGU--UGCCACCAGAGCUGC-GACUGCA--------------------CACGCCCAAAU-----AUAA-AAUUUUGUAGUAAUUACAUAUUUGCUC ..............((....-.(((((--(((..........))-)))))).--------------------....))(((((-----((..-((((.......))))..)))))))... ( -16.30) >DroYak_CAF1 41463 92 - 1 AAAAUAUAGAAAACGGAUUU-UGUAGU-UUGCCAUCAGAGCUGC-GGCUGCA--------------------CACGGCCAAAU-----AUAAAAAUUUUUUAGUAAUUACAUGUUUGCAC ......(((((((.......-.(((((-((.......)))))))-(((((..--------------------..)))))....-----.......)))))))((((........)))).. ( -18.30) >DroPer_CAF1 87542 112 - 1 AAAAUAUAGAAAACAGAUUUUUGUAGU-UCGCCGCCUCCGCCGC-AGACGCAGACGCAGUCACUGCCACUGCCACUGCCAAAC-----ACGC-AUUUUUUUAGUAAUUACAUAUUUGCAC .............(((((...((((((-((((.((....)).))-....((((..(((((.......)))))..)))).....-----....-.........).))))))).)))))... ( -21.90) >consensus AAAAUAUAGAAAACGGAUUU_UGCAGU_UUGCCAUCAGAGCUGC_GGCUGCA____________________CACGGCCAAAU_____AUAA_AAUUUUUUAGUAAUUACAUAUUUGCAC ......(((((((.........(((((.(((....))).))))).(((((........................)))))................)))))))((((........)))).. ( -7.68 = -9.44 + 1.76)

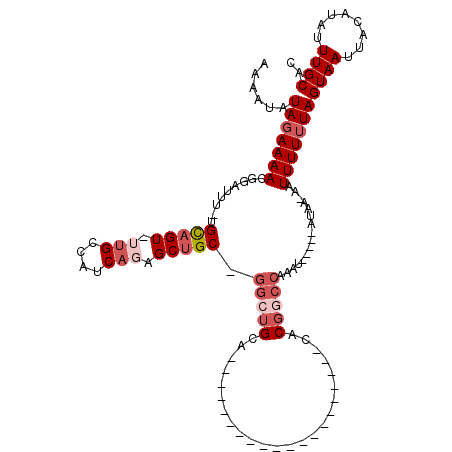
| Location | 15,122,293 – 15,122,389 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -7.18 |
| Energy contribution | -6.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15122293 96 - 22224390 CAUU---UGUGGGGCCCUUUGUGGGGCCUCAUUAUGCUGGAAAAUAUAGAAAACGGAUUU-UGCAGUUUUGCCAUCAGAGCUGCUGGCUGUA--------------------CACGGCCA (((.---.((((((((((....)))))))))).)))(((..............)))....-.(((((((((....))))))))).(((((..--------------------..))))). ( -37.94) >DroSec_CAF1 33953 91 - 1 CAUU---UGU---GCCCUUUGUGGGGCCUCAUUAUGCUG-AAAAUAUAGAAAACGGAUUU-UGCAGUUUUGCCAUCAGAGCUGC-GGCUGUA--------------------CACGGCCA .(((---(((---(((((....))))).(((......))-)...........))))))..-.(((((((((....)))))))))-(((((..--------------------..))))). ( -30.00) >DroEre_CAF1 40906 89 - 1 CAGU---GGG---GCCCUUUGUGA-GCCUCAUUAUGCUGGAAAAUAUAGAAAACGGAUUU-CGCAGU--UGCCACCAGAGCUGC-GACUGCA--------------------CACGCCCA .(((---(((---((.(.....).-))))))))..((((.........((((.....)))-)(((((--(((..........))-)))))).--------------------)).))... ( -25.80) >DroYak_CAF1 41498 90 - 1 CCGU---UGG---GCCCUUUGUGA-GCCUCAUUAUGCUGGAAAAUAUAGAAAACGGAUUU-UGUAGU-UUGCCAUCAGAGCUGC-GGCUGCA--------------------CACGGCCA ....---..(---(((...(((((-(((.((...((.((((((.(((((((......)))-)))).)-)).))).))....)).-)))).))--------------------)).)))). ( -27.70) >DroPer_CAF1 87576 117 - 1 CCGUCUCUUUACGAUCCUUUGUGG-GUCUCAUUAUGGUGAAAAAUAUAGAAAACAGAUUUUUGUAGU-UCGCCGCCUCCGCCGC-AGACGCAGACGCAGUCACUGCCACUGCCACUGCCA ..(((((.....(((((.....))-))).......((((((...((((((((.....)))))))).)-))))).........).-))))((((..(((((.......)))))..)))).. ( -33.60) >consensus CAGU___UGU___GCCCUUUGUGG_GCCUCAUUAUGCUGGAAAAUAUAGAAAACGGAUUU_UGCAGU_UUGCCAUCAGAGCUGC_GGCUGCA____________________CACGGCCA ....................(((........(((((.(....).))))).............(((((...((.......)).....))))).....................)))..... ( -7.18 = -6.94 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:10 2006