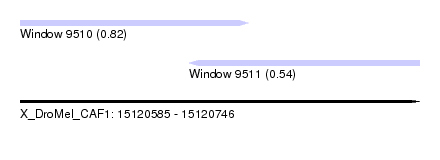
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 15,120,585 – 15,120,746 |
| Length | 161 |
| Max. P | 0.816839 |

| Location | 15,120,585 – 15,120,677 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -14.38 |
| Consensus MFE | -1.38 |
| Energy contribution | -1.66 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.10 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15120585 92 + 22224390 AUGACACUUUUGGCAAUUUUCCAGCCGCAUUUAUAUUUGCGUAAUUUCAUUU-----ACAAGCACAAAAACACUCAUAU----AC---ACAUAA---------AUUAAUUUAU ((((...(((((((............(((........)))((((......))-----))..)).)))))....))))..----..---......---------.......... ( -11.80) >DroSec_CAF1 32260 92 + 1 AUGACACUUUUGGCAAUUUUCCAGCCGCAUUUAUAUUUGCGUAAUUUCAUUU-----ACAAGCACAAAAACACUCAUAU----AC---ACAUAA---------AUUAAUUUAU ((((...(((((((............(((........)))((((......))-----))..)).)))))....))))..----..---......---------.......... ( -11.80) >DroEre_CAF1 39290 92 + 1 AUGACACUUUUGGCAAUUUUCCAGCCGCAUUUAUAUUUGCGUAAUUUCAUUU-----ACAAGCACAAACAAACGCAUAC----AC---ACACAA---------AUUCAUUUAU (((.....((((((............(((........)))((((......))-----))..)).))))......)))..----..---......---------.......... ( -9.90) >DroYak_CAF1 39781 92 + 1 AUGACACUUUUGGCAAUUUUCCAACCGCUUUUAUAUUUGCGUAAUUUCAUUU-----ACAAGCACAAAAGCACUUAUAU----AC---ACAUAA---------AUUAAUUUAU ......((((((((............((..........))((((......))-----))..)).)))))).........----..---......---------.......... ( -11.00) >DroPer_CAF1 86810 112 + 1 AUGACACUU-UGACCAUUUUGCAGCUGCAUUCACGCGGAUGUAAUCCCAUCUGAACAAAAAGCACACGAAAGCAUAUGUCGGUGCACUGCAUAAAUGUUAUUUAUUCAUUUAU ((((.....-((((.(((((((((.(((((..(((((((((......))))))........((........))...)))..)))))))))).)))))))).....)))).... ( -27.40) >consensus AUGACACUUUUGGCAAUUUUCCAGCCGCAUUUAUAUUUGCGUAAUUUCAUUU_____ACAAGCACAAAAACACUCAUAU____AC___ACAUAA_________AUUAAUUUAU .........................((((........))))........................................................................ ( -1.38 = -1.66 + 0.28)

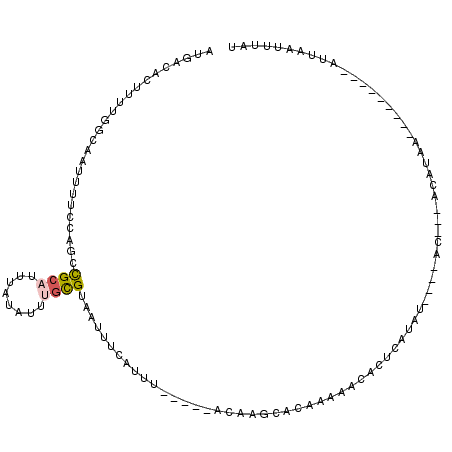
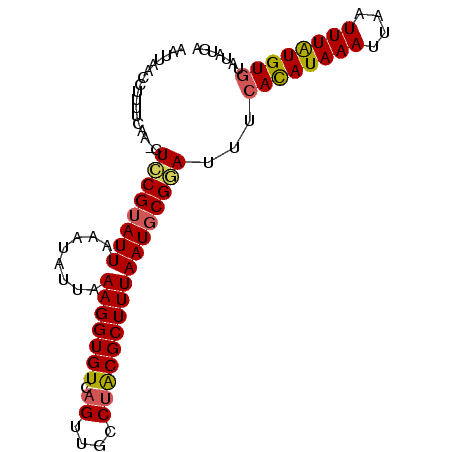
| Location | 15,120,653 – 15,120,746 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -17.88 |
| Consensus MFE | -14.11 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15120653 93 - 22224390 AAUUAACCUUUUCAA-CUCCGUAUUAAAUAUUAAAGGUGUCAGUUCGCUACGCUUUAAUGCGGAUUUCACAUAAAUUAAUUUAUGUGUAUAUGA ...........(((.-.((((((((((........(((((.((....)))))))))))))))))...((((((((....))))))))....))) ( -20.90) >DroSec_CAF1 32328 94 - 1 AAUUAACCUUUUCAAGCUUCGUAUUAAAUAUAAAAGGUGUCAGUUGUCUACGCUUUAAUGCGAAUUUCACAUAAAUUAAUUUAUGUGUAUAUGA ...........(((...((((((((((........(((((.((....)))))))))))))))))...((((((((....))))))))....))) ( -17.80) >DroEre_CAF1 39358 93 - 1 AAUUAAACUUUUCAA-CUCCGUAUUAAAUAUUAAAGGUGUCAGUUGCCAACGCUUUAAUGCGGAUUUAACAUAAAUGAAUUUGUGUGUGUAUGC ...............-.(((((((((((.......((((.....)))).....)))))))))))....(((((((....)))))))........ ( -17.20) >DroYak_CAF1 39849 93 - 1 AAUUAACCUUUUGCA-CUACGAAUUUAAGUUUAAAGGUGUCAGAUGCCUGCGCUUUAAUGCGGAUUUCAUAUAAAUUAAUUUAUGUGUAUAUAA .........((((((-....((((....)))).((((((.(((....)))))))))..))))))...((((((((....))))))))....... ( -15.60) >consensus AAUUAACCUUUUCAA_CUCCGUAUUAAAUAUUAAAGGUGUCAGUUGCCUACGCUUUAAUGCGGAUUUCACAUAAAUUAAUUUAUGUGUAUAUGA .................((((((((........(((((((.((....)))))))))))))))))...((((((((....))))))))....... (-14.11 = -14.30 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:08 2006