| Sequence ID | X_DroMel_CAF1 |
|---|---|
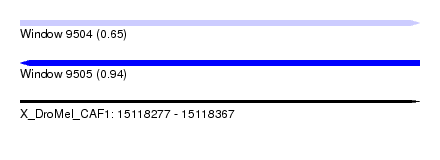
| Location | 15,118,277 – 15,118,367 |
| Length | 90 |
| Max. P | 0.942991 |

| Location | 15,118,277 – 15,118,367 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -18.14 |
| Consensus MFE | -14.43 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15118277 90 + 22224390 UAUGCCACGAAAAGUAUGCACAAGUUUGUGCCAAUCCUUCCUAAA----AAAAUCCCAGCCAAACUGGGUGGUAUGGGCAAAA-AAUAGAAAAAA (((((((((((......(((((....)))))......))).....----......((((.....)))))))))))).......-........... ( -20.70) >DroSec_CAF1 29990 95 + 1 UAUGCCACGAAAAGUAUGCACAAGUUUGUGCCAAUCCUCCCUAAAAAAAAAAAUCCCAGCCAAGCUUGGUGGUAUAGGCAAAGAAAGAAAAAUAA ((((((((.....(....).((((((((.((...........................))))))))))))))))))................... ( -18.73) >DroEre_CAF1 37152 87 + 1 UAUGCCACGAGAAGUAUGCACAAGUUUUUGCCAAUCCUCCCUAAA----AAAAUCCCAGCCAAGCCUGGUGGUAUAGGCAAAA-AAUAAAAA--- ..((((..(((......(((........))).....)))......----...(((((((......)))).)))...))))...-........--- ( -15.00) >consensus UAUGCCACGAAAAGUAUGCACAAGUUUGUGCCAAUCCUCCCUAAA____AAAAUCCCAGCCAAGCUUGGUGGUAUAGGCAAAA_AAUAAAAA_AA ..((((...........(((((....)))))........................(((.((......)))))....))))............... (-14.43 = -14.77 + 0.33)

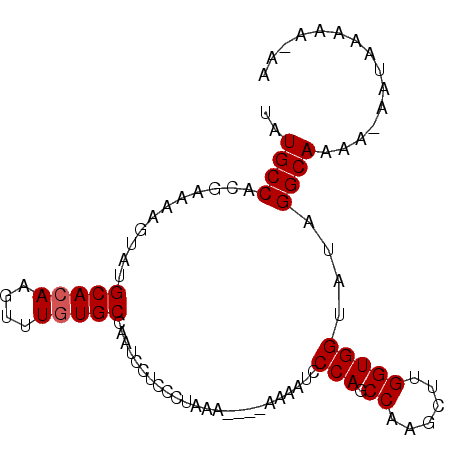
| Location | 15,118,277 – 15,118,367 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -23.23 |
| Consensus MFE | -21.04 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 15118277 90 - 22224390 UUUUUUCUAUU-UUUUGCCCAUACCACCCAGUUUGGCUGGGAUUUU----UUUAGGAAGGAUUGGCACAAACUUGUGCAUACUUUUCGUGGCAUA ...........-...(((((......(((((.....))))).....----....((((((....(((((....)))))...))))))).)))).. ( -26.20) >DroSec_CAF1 29990 95 - 1 UUAUUUUUCUUUCUUUGCCUAUACCACCAAGCUUGGCUGGGAUUUUUUUUUUUAGGGAGGAUUGGCACAAACUUGUGCAUACUUUUCGUGGCAUA ...............((((....((((((....))).)))..............(..(((....(((((....)))))...)))..)..)))).. ( -22.40) >DroEre_CAF1 37152 87 - 1 ---UUUUUAUU-UUUUGCCUAUACCACCAGGCUUGGCUGGGAUUUU----UUUAGGGAGGAUUGGCAAAAACUUGUGCAUACUUCUCGUGGCAUA ---.......(-(((((((.......((((......))))((((((----(.....)))))))))))))))..(((((.(((.....)))))))) ( -21.10) >consensus UU_UUUUUAUU_UUUUGCCUAUACCACCAAGCUUGGCUGGGAUUUU____UUUAGGGAGGAUUGGCACAAACUUGUGCAUACUUUUCGUGGCAUA ...............((((....(((((......)).)))..............((((((....(((((....)))))...))))))..)))).. (-21.04 = -20.93 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:03 2006